Gene
KWMTBOMO04427 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005409
Annotation
PREDICTED:_signal_recognition_particle_receptor_beta_subunit_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.23 PlasmaMembrane Reliability : 1.43
Sequence
CDS
ATGGAATCTGAAGTGGTTAAAGAAAAGATTCATACAGAACCGATCCTGAACGATCCACGATACATAACCTTACTTAGCCTTATTGTTCTAGCAGTAACCCTCATATTTTGGTGGATTTTCTCTCGAAGGTATACTCTTCGCAACTCAGTATTGTTAATGGGCCTATCAGACTCTGGCAAGACTTTGCTGTTTGTAAGACTCGCTTACTCTCAATACAGACAGACTTTTACTTCTATGAAAGAAAATATTGAAGAGTATATAACTTCTAATAAAACATTGAAAATAGTTGATCTGCCTGGACAAGAGAGACTGAGAAATAAATTCTTTGAACAGCACAAGAGTAGTGCTAAGGGAATTGTGTTTGTGATTGATTCTATCAATATACAGAAAGAAATCAGAGATGTAGCAGAATACCTATACACAATCTTGTGTGATCCGATCATTCAGGGAAACACAACTCCCCTTCTAATATTGTGCAACAAACAGGACCAGCCATTAGCAAAAGGAAGCCAAGTTATCAAAGGATTACTCGAAAAAGAAATAAATTTAGTTCGTGTGACCAAATCGAATCAACTGCAGTCCGTCGATCCGTCTGAAGGCAACATCGGGTCGTTCCTAGGTAAAGAGGGTAAAGATTTCGAATTCTCTCACATTAGAAATAAAGTCGAATTCGCAGAGTGCTCGGCCAACACGAATGACGATGAAAATCCGACTGACATCAAGCCTCTGCAGGATTGGATCTCGAAGCTGTAG
Protein
MESEVVKEKIHTEPILNDPRYITLLSLIVLAVTLIFWWIFSRRYTLRNSVLLMGLSDSGKTLLFVRLAYSQYRQTFTSMKENIEEYITSNKTLKIVDLPGQERLRNKFFEQHKSSAKGIVFVIDSINIQKEIRDVAEYLYTILCDPIIQGNTTPLLILCNKQDQPLAKGSQVIKGLLEKEINLVRVTKSNQLQSVDPSEGNIGSFLGKEGKDFEFSHIRNKVEFAECSANTNDDENPTDIKPLQDWISKL
Summary
Uniprot
H9J7B7
Q0N2R6
I4DJS1
A0A2A4IZT5
A0A212EI17
A0A194RBP0
+ More
S4NYF6 A0A1E1WR94 A0A2H1WB90 A0A154PRJ6 A0A0J7P4Z5 A0A232FAT8 E2AUT6 K7IX44 A0A151WWI8 A0A195D7I8 E2BYA5 A0A088A8V7 A0A2A3EIM1 V9IGH1 A0A0M8ZV53 A0A158NNH1 F4WQH2 E0VFZ7 A0A195F6Z7 A0A2P8XMD1 A0A1L8DVQ5 A0A195DKN0 A0A2J7QR38 A0A0L7R3R7 A0A1B0D5W5 A0A026W8U0 T1IJD7 A0A1B6EEG2 A0A1Q3FWQ9 A0A0K8TSV2 A0A1Q3G1Z6 A0A1B6E0T6 A0A1S3DRH5 A0A067QGT4 A0A0P5TWP1 A0A023EKZ5 D6X187 A0A1S4FNN0 A0A034WBQ9 Q16UQ1 A0A0P5T3G0 A0A0K8V0M7 A0A0P4ZW46 A0A1S3D7Q3 A0A0P5BQL2 A0A0P5KBS0 A0A0A1WDW3 A0A0P5AIY1 A0A0P4YNH9 A0A1B1UZL3 A0A3S1HC35 A0A0P5KQH7 A0A0P6H7V8 A0A0P5X800 A0A0P6EWC5 A0A0P5PQT5 B5M0T7 A0A1Y1LVG5 A0A336LPP0 B4KZ72 A0A0P5H715 A0A069DR19 A0A0N8CBZ4 A0A0P6GWW0 A0A224XWH5 E9FSV7 A0A2M3ZL65 A0A1U7RVM5 A0A1B6EHE8 A0A3M6T4X4 A0A2M4AHE4 A0A2M3Z7P2 Q9VSN9 A0A224Z8M7 A0A182FFR9 Q6IDG2 A0A0M4ED82 A0A182R2F4 A0A2M4BY06 W5JAN6 A0A1B6K7G5 A0A2M4AIA6 A0A2M3ZKZ1 A0A2M4BY78 A0A0P4ZYV2 B4PFK3 B3M673 A0A087UR91 A0A182Q9H6 A0A2L2YLZ9 A0A131XE64 A0A0P4W032
S4NYF6 A0A1E1WR94 A0A2H1WB90 A0A154PRJ6 A0A0J7P4Z5 A0A232FAT8 E2AUT6 K7IX44 A0A151WWI8 A0A195D7I8 E2BYA5 A0A088A8V7 A0A2A3EIM1 V9IGH1 A0A0M8ZV53 A0A158NNH1 F4WQH2 E0VFZ7 A0A195F6Z7 A0A2P8XMD1 A0A1L8DVQ5 A0A195DKN0 A0A2J7QR38 A0A0L7R3R7 A0A1B0D5W5 A0A026W8U0 T1IJD7 A0A1B6EEG2 A0A1Q3FWQ9 A0A0K8TSV2 A0A1Q3G1Z6 A0A1B6E0T6 A0A1S3DRH5 A0A067QGT4 A0A0P5TWP1 A0A023EKZ5 D6X187 A0A1S4FNN0 A0A034WBQ9 Q16UQ1 A0A0P5T3G0 A0A0K8V0M7 A0A0P4ZW46 A0A1S3D7Q3 A0A0P5BQL2 A0A0P5KBS0 A0A0A1WDW3 A0A0P5AIY1 A0A0P4YNH9 A0A1B1UZL3 A0A3S1HC35 A0A0P5KQH7 A0A0P6H7V8 A0A0P5X800 A0A0P6EWC5 A0A0P5PQT5 B5M0T7 A0A1Y1LVG5 A0A336LPP0 B4KZ72 A0A0P5H715 A0A069DR19 A0A0N8CBZ4 A0A0P6GWW0 A0A224XWH5 E9FSV7 A0A2M3ZL65 A0A1U7RVM5 A0A1B6EHE8 A0A3M6T4X4 A0A2M4AHE4 A0A2M3Z7P2 Q9VSN9 A0A224Z8M7 A0A182FFR9 Q6IDG2 A0A0M4ED82 A0A182R2F4 A0A2M4BY06 W5JAN6 A0A1B6K7G5 A0A2M4AIA6 A0A2M3ZKZ1 A0A2M4BY78 A0A0P4ZYV2 B4PFK3 B3M673 A0A087UR91 A0A182Q9H6 A0A2L2YLZ9 A0A131XE64 A0A0P4W032
Pubmed
19121390
22651552
26354079
22118469
23622113
28648823
+ More
20798317 20075255 21347285 21719571 20566863 29403074 24508170 30249741 26369729 24845553 24945155 18362917 19820115 25348373 17510324 25830018 19166301 28004739 17994087 26334808 21292972 30382153 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28797301 20920257 23761445 17550304 26561354 28049606 27129103
20798317 20075255 21347285 21719571 20566863 29403074 24508170 30249741 26369729 24845553 24945155 18362917 19820115 25348373 17510324 25830018 19166301 28004739 17994087 26334808 21292972 30382153 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28797301 20920257 23761445 17550304 26561354 28049606 27129103
EMBL
BABH01013609
DQ646415
ABH10805.1
AK401539
KQ459589
BAM18161.1
+ More
KPI97689.1 NWSH01004148 PCG65427.1 AGBW02014758 OWR41110.1 KQ460397 KPJ15253.1 GAIX01013825 JAA78735.1 GDQN01001509 JAT89545.1 ODYU01007502 SOQ50328.1 KQ435087 KZC14536.1 LBMM01000096 KMR04984.1 NNAY01000601 OXU27447.1 GL442888 EFN62811.1 AAZX01000571 AAZX01006238 KQ982691 KYQ52253.1 KQ976749 KYN08817.1 GL451420 EFN79384.1 KZ288230 PBC31643.1 JR042163 AEY59566.1 KQ435855 KOX70646.1 ADTU01021600 GL888273 EGI63572.1 DS235129 EEB12303.1 KQ981744 KYN36223.1 PYGN01001735 PSN33114.1 GFDF01003591 JAV10493.1 KQ980765 KYN13406.1 NEVH01011988 PNF31046.1 KQ414661 KOC65535.1 AJVK01000417 KK107343 QOIP01000013 EZA52398.1 RLU15481.1 AFFK01020433 JH430253 GEDC01000959 JAS36339.1 GFDL01003028 JAV32017.1 GDAI01000390 JAI17213.1 GFDL01001210 JAV33835.1 GEDC01005801 JAS31497.1 KK853448 KDR07565.1 GDIP01121558 JAL82156.1 GAPW01003746 JAC09852.1 KQ971372 EFA10591.1 GAKP01005927 GAKP01005926 JAC53026.1 CH477616 EAT38257.1 GDIP01136061 JAL67653.1 GDHF01019913 JAI32401.1 GDIP01220000 GDIP01207948 GDIP01127962 LRGB01003123 JAJ15454.1 KZS04441.1 GDIP01182147 JAJ41255.1 GDIQ01244163 GDIQ01192831 GDIQ01173879 GDIQ01143795 GDIQ01031433 JAK58894.1 GBXI01017195 JAC97096.1 GDIP01198354 JAJ25048.1 GDIP01225099 JAI98302.1 KU568374 ANW09661.1 RQTK01000673 RUS76303.1 GDIQ01180732 JAK70993.1 GDIQ01212716 GDIQ01131602 GDIQ01042487 GDIQ01032936 JAN61801.1 GDIP01076113 JAM27602.1 GDIQ01057179 JAN37558.1 GDIQ01135551 JAL16175.1 EU930273 ACH56901.1 GEZM01050414 GEZM01050413 JAV75596.1 UFQT01000102 SSX20022.1 CH933809 EDW17869.1 GDIQ01230318 JAK21407.1 GBGD01002733 JAC86156.1 GDIQ01094707 JAL57019.1 GDIQ01027371 JAN67366.1 GFTR01003933 JAW12493.1 GL732524 EFX89260.1 GGFM01008449 MBW29200.1 GECZ01032422 JAS37347.1 RCHS01004337 RMX35930.1 GGFK01006821 MBW40142.1 GGFM01003781 MBW24532.1 AF181658 AE014296 AAD55443.1 AAF50377.2 AGB94291.1 GFPF01011758 MAA22904.1 BT014643 AAT27267.1 CP012525 ALC43508.1 GGFJ01008824 MBW57965.1 ADMH02001932 ETN60463.1 GECU01000308 JAT07399.1 GGFK01007141 MBW40462.1 GGFM01008448 MBW29199.1 GGFJ01008823 MBW57964.1 GDIP01209454 JAJ13948.1 CM000159 EDW93129.1 CH902618 EDV39693.2 KK121162 KFM79880.1 AXCN02000584 IAAA01039247 LAA09148.1 GEFH01003939 JAP64642.1 GDKW01001478 JAI55117.1
KPI97689.1 NWSH01004148 PCG65427.1 AGBW02014758 OWR41110.1 KQ460397 KPJ15253.1 GAIX01013825 JAA78735.1 GDQN01001509 JAT89545.1 ODYU01007502 SOQ50328.1 KQ435087 KZC14536.1 LBMM01000096 KMR04984.1 NNAY01000601 OXU27447.1 GL442888 EFN62811.1 AAZX01000571 AAZX01006238 KQ982691 KYQ52253.1 KQ976749 KYN08817.1 GL451420 EFN79384.1 KZ288230 PBC31643.1 JR042163 AEY59566.1 KQ435855 KOX70646.1 ADTU01021600 GL888273 EGI63572.1 DS235129 EEB12303.1 KQ981744 KYN36223.1 PYGN01001735 PSN33114.1 GFDF01003591 JAV10493.1 KQ980765 KYN13406.1 NEVH01011988 PNF31046.1 KQ414661 KOC65535.1 AJVK01000417 KK107343 QOIP01000013 EZA52398.1 RLU15481.1 AFFK01020433 JH430253 GEDC01000959 JAS36339.1 GFDL01003028 JAV32017.1 GDAI01000390 JAI17213.1 GFDL01001210 JAV33835.1 GEDC01005801 JAS31497.1 KK853448 KDR07565.1 GDIP01121558 JAL82156.1 GAPW01003746 JAC09852.1 KQ971372 EFA10591.1 GAKP01005927 GAKP01005926 JAC53026.1 CH477616 EAT38257.1 GDIP01136061 JAL67653.1 GDHF01019913 JAI32401.1 GDIP01220000 GDIP01207948 GDIP01127962 LRGB01003123 JAJ15454.1 KZS04441.1 GDIP01182147 JAJ41255.1 GDIQ01244163 GDIQ01192831 GDIQ01173879 GDIQ01143795 GDIQ01031433 JAK58894.1 GBXI01017195 JAC97096.1 GDIP01198354 JAJ25048.1 GDIP01225099 JAI98302.1 KU568374 ANW09661.1 RQTK01000673 RUS76303.1 GDIQ01180732 JAK70993.1 GDIQ01212716 GDIQ01131602 GDIQ01042487 GDIQ01032936 JAN61801.1 GDIP01076113 JAM27602.1 GDIQ01057179 JAN37558.1 GDIQ01135551 JAL16175.1 EU930273 ACH56901.1 GEZM01050414 GEZM01050413 JAV75596.1 UFQT01000102 SSX20022.1 CH933809 EDW17869.1 GDIQ01230318 JAK21407.1 GBGD01002733 JAC86156.1 GDIQ01094707 JAL57019.1 GDIQ01027371 JAN67366.1 GFTR01003933 JAW12493.1 GL732524 EFX89260.1 GGFM01008449 MBW29200.1 GECZ01032422 JAS37347.1 RCHS01004337 RMX35930.1 GGFK01006821 MBW40142.1 GGFM01003781 MBW24532.1 AF181658 AE014296 AAD55443.1 AAF50377.2 AGB94291.1 GFPF01011758 MAA22904.1 BT014643 AAT27267.1 CP012525 ALC43508.1 GGFJ01008824 MBW57965.1 ADMH02001932 ETN60463.1 GECU01000308 JAT07399.1 GGFK01007141 MBW40462.1 GGFM01008448 MBW29199.1 GGFJ01008823 MBW57964.1 GDIP01209454 JAJ13948.1 CM000159 EDW93129.1 CH902618 EDV39693.2 KK121162 KFM79880.1 AXCN02000584 IAAA01039247 LAA09148.1 GEFH01003939 JAP64642.1 GDKW01001478 JAI55117.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000053240
UP000076502
+ More
UP000036403 UP000215335 UP000000311 UP000002358 UP000075809 UP000078542 UP000008237 UP000005203 UP000242457 UP000053105 UP000005205 UP000007755 UP000009046 UP000078541 UP000245037 UP000078492 UP000235965 UP000053825 UP000092462 UP000053097 UP000279307 UP000079169 UP000027135 UP000007266 UP000008820 UP000076858 UP000271974 UP000009192 UP000000305 UP000189705 UP000275408 UP000000803 UP000069272 UP000092553 UP000075900 UP000000673 UP000002282 UP000007801 UP000054359 UP000075886
UP000036403 UP000215335 UP000000311 UP000002358 UP000075809 UP000078542 UP000008237 UP000005203 UP000242457 UP000053105 UP000005205 UP000007755 UP000009046 UP000078541 UP000245037 UP000078492 UP000235965 UP000053825 UP000092462 UP000053097 UP000279307 UP000079169 UP000027135 UP000007266 UP000008820 UP000076858 UP000271974 UP000009192 UP000000305 UP000189705 UP000275408 UP000000803 UP000069272 UP000092553 UP000075900 UP000000673 UP000002282 UP000007801 UP000054359 UP000075886
Pfam
PF09439 SRPRB
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
H9J7B7
Q0N2R6
I4DJS1
A0A2A4IZT5
A0A212EI17
A0A194RBP0
+ More
S4NYF6 A0A1E1WR94 A0A2H1WB90 A0A154PRJ6 A0A0J7P4Z5 A0A232FAT8 E2AUT6 K7IX44 A0A151WWI8 A0A195D7I8 E2BYA5 A0A088A8V7 A0A2A3EIM1 V9IGH1 A0A0M8ZV53 A0A158NNH1 F4WQH2 E0VFZ7 A0A195F6Z7 A0A2P8XMD1 A0A1L8DVQ5 A0A195DKN0 A0A2J7QR38 A0A0L7R3R7 A0A1B0D5W5 A0A026W8U0 T1IJD7 A0A1B6EEG2 A0A1Q3FWQ9 A0A0K8TSV2 A0A1Q3G1Z6 A0A1B6E0T6 A0A1S3DRH5 A0A067QGT4 A0A0P5TWP1 A0A023EKZ5 D6X187 A0A1S4FNN0 A0A034WBQ9 Q16UQ1 A0A0P5T3G0 A0A0K8V0M7 A0A0P4ZW46 A0A1S3D7Q3 A0A0P5BQL2 A0A0P5KBS0 A0A0A1WDW3 A0A0P5AIY1 A0A0P4YNH9 A0A1B1UZL3 A0A3S1HC35 A0A0P5KQH7 A0A0P6H7V8 A0A0P5X800 A0A0P6EWC5 A0A0P5PQT5 B5M0T7 A0A1Y1LVG5 A0A336LPP0 B4KZ72 A0A0P5H715 A0A069DR19 A0A0N8CBZ4 A0A0P6GWW0 A0A224XWH5 E9FSV7 A0A2M3ZL65 A0A1U7RVM5 A0A1B6EHE8 A0A3M6T4X4 A0A2M4AHE4 A0A2M3Z7P2 Q9VSN9 A0A224Z8M7 A0A182FFR9 Q6IDG2 A0A0M4ED82 A0A182R2F4 A0A2M4BY06 W5JAN6 A0A1B6K7G5 A0A2M4AIA6 A0A2M3ZKZ1 A0A2M4BY78 A0A0P4ZYV2 B4PFK3 B3M673 A0A087UR91 A0A182Q9H6 A0A2L2YLZ9 A0A131XE64 A0A0P4W032
S4NYF6 A0A1E1WR94 A0A2H1WB90 A0A154PRJ6 A0A0J7P4Z5 A0A232FAT8 E2AUT6 K7IX44 A0A151WWI8 A0A195D7I8 E2BYA5 A0A088A8V7 A0A2A3EIM1 V9IGH1 A0A0M8ZV53 A0A158NNH1 F4WQH2 E0VFZ7 A0A195F6Z7 A0A2P8XMD1 A0A1L8DVQ5 A0A195DKN0 A0A2J7QR38 A0A0L7R3R7 A0A1B0D5W5 A0A026W8U0 T1IJD7 A0A1B6EEG2 A0A1Q3FWQ9 A0A0K8TSV2 A0A1Q3G1Z6 A0A1B6E0T6 A0A1S3DRH5 A0A067QGT4 A0A0P5TWP1 A0A023EKZ5 D6X187 A0A1S4FNN0 A0A034WBQ9 Q16UQ1 A0A0P5T3G0 A0A0K8V0M7 A0A0P4ZW46 A0A1S3D7Q3 A0A0P5BQL2 A0A0P5KBS0 A0A0A1WDW3 A0A0P5AIY1 A0A0P4YNH9 A0A1B1UZL3 A0A3S1HC35 A0A0P5KQH7 A0A0P6H7V8 A0A0P5X800 A0A0P6EWC5 A0A0P5PQT5 B5M0T7 A0A1Y1LVG5 A0A336LPP0 B4KZ72 A0A0P5H715 A0A069DR19 A0A0N8CBZ4 A0A0P6GWW0 A0A224XWH5 E9FSV7 A0A2M3ZL65 A0A1U7RVM5 A0A1B6EHE8 A0A3M6T4X4 A0A2M4AHE4 A0A2M3Z7P2 Q9VSN9 A0A224Z8M7 A0A182FFR9 Q6IDG2 A0A0M4ED82 A0A182R2F4 A0A2M4BY06 W5JAN6 A0A1B6K7G5 A0A2M4AIA6 A0A2M3ZKZ1 A0A2M4BY78 A0A0P4ZYV2 B4PFK3 B3M673 A0A087UR91 A0A182Q9H6 A0A2L2YLZ9 A0A131XE64 A0A0P4W032
PDB
6FRK
E-value=1.96863e-45,
Score=457
Ontologies
GO
PANTHER
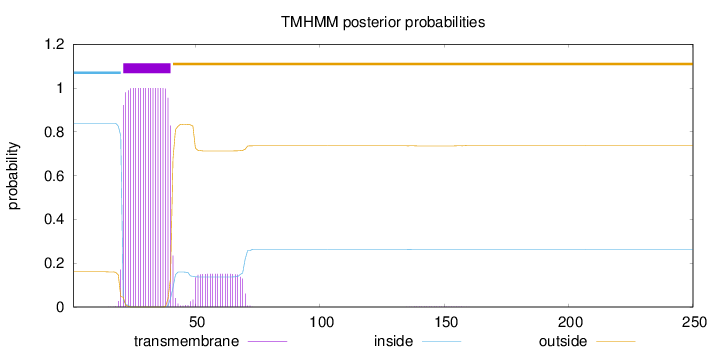
Topology
Length:
250
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.32326
Exp number, first 60 AAs:
21.8914
Total prob of N-in:
0.83937
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 40
outside
41 - 250
Population Genetic Test Statistics
Pi
228.998003
Theta
173.363383
Tajima's D
1.252166
CLR
13.979575
CSRT
0.726063696815159
Interpretation
Uncertain
