Gene
KWMTBOMO04425
Pre Gene Modal
BGIBMGA005408
Annotation
Histidine_protein_methyltransferase_1-like_protein_[Operophtera_brumata]
Full name
Histidine protein methyltransferase 1 homolog
Alternative Name
Methyltransferase-like protein 18
Location in the cell
Cytoplasmic Reliability : 2.875
Sequence
CDS
ATGACTGATTTTAGATTCAATTTTTCTGGAGCGGAGTCTGAAGTAGAACACAAAGACGATGAAGAAATTAGCTGGCTAGCCAGTGAAGAGGTTAAACCGGATAAACAAATAAAAGAAGTAGATGAAATAAATTTACTTTACCTTGCCTTACTGCCATGCCATGAAACGGTAATAAGACATATTATGCCAACTAAAATTATTGCAGAACTAAAAAATAAAGAATTACAAAATGTTTTGAATACAGCTGAAAAAGAACATAGCGATCTAGTCACAGGAAAATACGAAGGTGGACTAAAAATATGGGAATGCACTTACGATTTGTTACAATACCTGATGCATAACAAGGATATAATAGAATTTCAGGATAAAAATGTTTTAGACCTGGGATGTGGAGCAGGAATATTAGGAATATATGCACTGCAAAATGGAGCTAATGTTACATTTCAAGATTATAATAAAGAAGTCTTGGAGAATGTTACTATTCCCAATGTATTAGTAAATATAGAAGAAAAATTCAGAGAAAAAGAGATAAAAAGATATTGA
Protein
MTDFRFNFSGAESEVEHKDDEEISWLASEEVKPDKQIKEVDEINLLYLALLPCHETVIRHIMPTKIIAELKNKELQNVLNTAEKEHSDLVTGKYEGGLKIWECTYDLLQYLMHNKDIIEFQDKNVLDLGCGAGILGIYALQNGANVTFQDYNKEVLENVTIPNVLVNIEEKFREKEIKRY
Summary
Description
Probable histidine methyltransferase.
Subunit
Interacts with GRWD1, RPL3 and members of the heat shock protein 90 and 70 families; these proteins may possibly be methylation substrates for the enzyme.
Similarity
Belongs to the methyltransferase superfamily. METTL18 family.
Keywords
Complete proteome
Methyltransferase
Phosphoprotein
Reference proteome
S-adenosyl-L-methionine
Transferase
Feature
chain Histidine protein methyltransferase 1 homolog
Uniprot
H9J7B6
A0A2H1WW20
A0A0L7LJF0
S4PIM8
A0A212EI04
A0A3S2NIV7
+ More
A0A194PWJ0 A0A194RDA1 B4LT18 B4G6Z2 A0A3B0K1T2 A0A1I8MXC2 Q29LZ9 A0A0M4EEK6 A0A232FAJ4 A0A0L0BX58 A0A151J0G4 A0A1W4V5P3 B4KIL3 B4NXC7 B4JA62 B4Q918 B4I2U6 J3JYG2 U4U9P3 K7IX46 A0A088A8S7 A0A1A9WXL1 A0A026W349 B3NAP7 Q8SYR9 F4WMQ7 B3MMY7 Q9VQK8 W8BTD5 A0A195B434 A0A158NQ94 D6X186 A0A1I8PYU0 A0A195CUN9 A0A3M6UM17 A0A0A1WP08 E2AIC4 A0A195FQG8 B4MW17 A0A0L7R3V6 A0A139WBZ2 A0A0K8WK52 S9XMZ9 A0A1A9XEN3 A0A1B0G9K6 A0A1W4XUG7 A0A1B0C571 A0A1B0BAG8 Q2KIJ2 A0A067QHT1 L8I7P9 A0A2J7PBM2 A0A2J7PBL4 A0A3B3B9M3 E9FYX8 A0A2J7PBL1 A0A1A8GSH2 A0A1S3WMU3 A0A1A8FSN5 A0A3Q7TCM6 A0A2P4T8D5 L7MSA7 A0A1D5P5E6 A0A2U3WJD4 F1RSC8 I3MXY5
A0A194PWJ0 A0A194RDA1 B4LT18 B4G6Z2 A0A3B0K1T2 A0A1I8MXC2 Q29LZ9 A0A0M4EEK6 A0A232FAJ4 A0A0L0BX58 A0A151J0G4 A0A1W4V5P3 B4KIL3 B4NXC7 B4JA62 B4Q918 B4I2U6 J3JYG2 U4U9P3 K7IX46 A0A088A8S7 A0A1A9WXL1 A0A026W349 B3NAP7 Q8SYR9 F4WMQ7 B3MMY7 Q9VQK8 W8BTD5 A0A195B434 A0A158NQ94 D6X186 A0A1I8PYU0 A0A195CUN9 A0A3M6UM17 A0A0A1WP08 E2AIC4 A0A195FQG8 B4MW17 A0A0L7R3V6 A0A139WBZ2 A0A0K8WK52 S9XMZ9 A0A1A9XEN3 A0A1B0G9K6 A0A1W4XUG7 A0A1B0C571 A0A1B0BAG8 Q2KIJ2 A0A067QHT1 L8I7P9 A0A2J7PBM2 A0A2J7PBL4 A0A3B3B9M3 E9FYX8 A0A2J7PBL1 A0A1A8GSH2 A0A1S3WMU3 A0A1A8FSN5 A0A3Q7TCM6 A0A2P4T8D5 L7MSA7 A0A1D5P5E6 A0A2U3WJD4 F1RSC8 I3MXY5
Pubmed
19121390
26227816
23622113
22118469
26354079
17994087
+ More
25315136 15632085 28648823 26108605 17550304 22936249 22516182 23537049 20075255 24508170 30249741 21719571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 21347285 18362917 19820115 30382153 25830018 20798317 23149746 24845553 22751099 29451363 21292972 15592404
25315136 15632085 28648823 26108605 17550304 22936249 22516182 23537049 20075255 24508170 30249741 21719571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 21347285 18362917 19820115 30382153 25830018 20798317 23149746 24845553 22751099 29451363 21292972 15592404
EMBL
BABH01013608
ODYU01011099
SOQ56624.1
JTDY01000856
KOB75658.1
GAIX01000144
+ More
JAA92416.1 AGBW02014758 OWR41107.1 RSAL01000025 RVE52147.1 KQ459589 KPI97687.1 KQ460397 KPJ15255.1 CH940649 EDW63849.1 CH479180 EDW29191.1 OUUW01000004 SPP79556.1 CH379060 EAL33896.2 CP012523 ALC38605.1 NNAY01000601 OXU27453.1 JRES01001205 KNC24632.1 KQ980658 KYN14760.1 CH933807 EDW11356.1 CM000157 EDW87484.1 CH916368 EDW03736.1 CM000361 CM002910 EDX03557.1 KMY87806.1 CH480820 EDW54091.1 APGK01029461 BT128290 KB740735 AEE63248.1 ENN79136.1 KB632136 ERL89053.1 AAZX01000600 KK107455 QOIP01000010 EZA50448.1 RLU17342.1 CH954177 EDV57570.1 AY071354 AAL48976.1 GL888218 EGI64534.1 CH902620 EDV31012.1 AE014134 BT030812 AAF51158.2 ABV82194.1 GAMC01004113 JAC02443.1 KQ976618 KYM79253.1 ADTU01022976 KQ971372 EFA10806.2 KQ977279 KYN04237.1 RCHS01001219 RMX54682.1 GBXI01014057 JAD00235.1 GL439754 EFN66835.1 KQ981382 KYN42169.1 CH963857 EDW75887.1 KQ414661 KOC65533.1 KYB25430.1 GDHF01000771 JAI51543.1 KB016393 EPY89558.1 CCAG010009121 JXJN01025882 JXJN01010999 BC112618 KK853387 KDR07955.1 JH881963 ELR51564.1 NEVH01027081 PNF13719.1 PNF13718.1 GL732527 EFX87694.1 PNF13717.1 HAEC01005272 SBQ73349.1 HAEB01015336 SBQ61863.1 PPHD01005382 POI32629.1 HQ890279 AEB61434.1 AADN05000934 AEMK02000022 AGTP01015049
JAA92416.1 AGBW02014758 OWR41107.1 RSAL01000025 RVE52147.1 KQ459589 KPI97687.1 KQ460397 KPJ15255.1 CH940649 EDW63849.1 CH479180 EDW29191.1 OUUW01000004 SPP79556.1 CH379060 EAL33896.2 CP012523 ALC38605.1 NNAY01000601 OXU27453.1 JRES01001205 KNC24632.1 KQ980658 KYN14760.1 CH933807 EDW11356.1 CM000157 EDW87484.1 CH916368 EDW03736.1 CM000361 CM002910 EDX03557.1 KMY87806.1 CH480820 EDW54091.1 APGK01029461 BT128290 KB740735 AEE63248.1 ENN79136.1 KB632136 ERL89053.1 AAZX01000600 KK107455 QOIP01000010 EZA50448.1 RLU17342.1 CH954177 EDV57570.1 AY071354 AAL48976.1 GL888218 EGI64534.1 CH902620 EDV31012.1 AE014134 BT030812 AAF51158.2 ABV82194.1 GAMC01004113 JAC02443.1 KQ976618 KYM79253.1 ADTU01022976 KQ971372 EFA10806.2 KQ977279 KYN04237.1 RCHS01001219 RMX54682.1 GBXI01014057 JAD00235.1 GL439754 EFN66835.1 KQ981382 KYN42169.1 CH963857 EDW75887.1 KQ414661 KOC65533.1 KYB25430.1 GDHF01000771 JAI51543.1 KB016393 EPY89558.1 CCAG010009121 JXJN01025882 JXJN01010999 BC112618 KK853387 KDR07955.1 JH881963 ELR51564.1 NEVH01027081 PNF13719.1 PNF13718.1 GL732527 EFX87694.1 PNF13717.1 HAEC01005272 SBQ73349.1 HAEB01015336 SBQ61863.1 PPHD01005382 POI32629.1 HQ890279 AEB61434.1 AADN05000934 AEMK02000022 AGTP01015049
Proteomes
UP000005204
UP000037510
UP000007151
UP000283053
UP000053268
UP000053240
+ More
UP000008792 UP000008744 UP000268350 UP000095301 UP000001819 UP000092553 UP000215335 UP000037069 UP000078492 UP000192221 UP000009192 UP000002282 UP000001070 UP000000304 UP000001292 UP000019118 UP000030742 UP000002358 UP000005203 UP000091820 UP000053097 UP000279307 UP000008711 UP000007755 UP000007801 UP000000803 UP000078540 UP000005205 UP000007266 UP000095300 UP000078542 UP000275408 UP000000311 UP000078541 UP000007798 UP000053825 UP000092443 UP000092444 UP000192223 UP000092460 UP000009136 UP000027135 UP000235965 UP000261560 UP000000305 UP000079721 UP000286640 UP000000539 UP000245340 UP000008227 UP000005215
UP000008792 UP000008744 UP000268350 UP000095301 UP000001819 UP000092553 UP000215335 UP000037069 UP000078492 UP000192221 UP000009192 UP000002282 UP000001070 UP000000304 UP000001292 UP000019118 UP000030742 UP000002358 UP000005203 UP000091820 UP000053097 UP000279307 UP000008711 UP000007755 UP000007801 UP000000803 UP000078540 UP000005205 UP000007266 UP000095300 UP000078542 UP000275408 UP000000311 UP000078541 UP000007798 UP000053825 UP000092443 UP000092444 UP000192223 UP000092460 UP000009136 UP000027135 UP000235965 UP000261560 UP000000305 UP000079721 UP000286640 UP000000539 UP000245340 UP000008227 UP000005215
Interpro
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
H9J7B6
A0A2H1WW20
A0A0L7LJF0
S4PIM8
A0A212EI04
A0A3S2NIV7
+ More
A0A194PWJ0 A0A194RDA1 B4LT18 B4G6Z2 A0A3B0K1T2 A0A1I8MXC2 Q29LZ9 A0A0M4EEK6 A0A232FAJ4 A0A0L0BX58 A0A151J0G4 A0A1W4V5P3 B4KIL3 B4NXC7 B4JA62 B4Q918 B4I2U6 J3JYG2 U4U9P3 K7IX46 A0A088A8S7 A0A1A9WXL1 A0A026W349 B3NAP7 Q8SYR9 F4WMQ7 B3MMY7 Q9VQK8 W8BTD5 A0A195B434 A0A158NQ94 D6X186 A0A1I8PYU0 A0A195CUN9 A0A3M6UM17 A0A0A1WP08 E2AIC4 A0A195FQG8 B4MW17 A0A0L7R3V6 A0A139WBZ2 A0A0K8WK52 S9XMZ9 A0A1A9XEN3 A0A1B0G9K6 A0A1W4XUG7 A0A1B0C571 A0A1B0BAG8 Q2KIJ2 A0A067QHT1 L8I7P9 A0A2J7PBM2 A0A2J7PBL4 A0A3B3B9M3 E9FYX8 A0A2J7PBL1 A0A1A8GSH2 A0A1S3WMU3 A0A1A8FSN5 A0A3Q7TCM6 A0A2P4T8D5 L7MSA7 A0A1D5P5E6 A0A2U3WJD4 F1RSC8 I3MXY5
A0A194PWJ0 A0A194RDA1 B4LT18 B4G6Z2 A0A3B0K1T2 A0A1I8MXC2 Q29LZ9 A0A0M4EEK6 A0A232FAJ4 A0A0L0BX58 A0A151J0G4 A0A1W4V5P3 B4KIL3 B4NXC7 B4JA62 B4Q918 B4I2U6 J3JYG2 U4U9P3 K7IX46 A0A088A8S7 A0A1A9WXL1 A0A026W349 B3NAP7 Q8SYR9 F4WMQ7 B3MMY7 Q9VQK8 W8BTD5 A0A195B434 A0A158NQ94 D6X186 A0A1I8PYU0 A0A195CUN9 A0A3M6UM17 A0A0A1WP08 E2AIC4 A0A195FQG8 B4MW17 A0A0L7R3V6 A0A139WBZ2 A0A0K8WK52 S9XMZ9 A0A1A9XEN3 A0A1B0G9K6 A0A1W4XUG7 A0A1B0C571 A0A1B0BAG8 Q2KIJ2 A0A067QHT1 L8I7P9 A0A2J7PBM2 A0A2J7PBL4 A0A3B3B9M3 E9FYX8 A0A2J7PBL1 A0A1A8GSH2 A0A1S3WMU3 A0A1A8FSN5 A0A3Q7TCM6 A0A2P4T8D5 L7MSA7 A0A1D5P5E6 A0A2U3WJD4 F1RSC8 I3MXY5
PDB
4RFQ
E-value=3.02993e-19,
Score=229
Ontologies
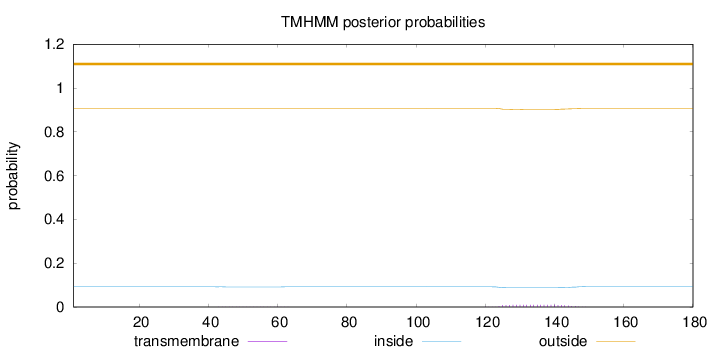
Topology
Length:
180
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.21059
Exp number, first 60 AAs:
0.01156
Total prob of N-in:
0.09226
outside
1 - 180
Population Genetic Test Statistics
Pi
126.831311
Theta
122.0665
Tajima's D
-0.323613
CLR
20.491889
CSRT
0.283685815709215
Interpretation
Uncertain
