Gene
KWMTBOMO04414
Annotation
gustatory_receptor_68_[Bombyx_mori]
Full name
Gustatory receptor
+ More
Gustatory receptor 68a
Gustatory receptor 68a
Location in the cell
PlasmaMembrane Reliability : 3.766
Sequence
CDS
ATGCCGTTATCGTTGAAACAACTGGAATTGATTTACATAAAGGCTTTTGAACTGAAAAGAGATATCAATAAAGCTTTTGAAGCTCCAATATTGTTGACTACAATGCAATGTTTTCACTCAATAGTAAGCGAATCACATATCATCTACCATGGTGCGGTAATGGAGCCACACATGGTACTACATTCAATAATGAATTGCTCTGTTTGGATACTATATCAGCTATTCAAATTATATATATTAGCTTCAACAGGTCATTTGCTACAGGAGAAGATACAACATTTTTCAAATCTAATACACTTCCATGGAAAAGGATTAACCGTGTATGGATTATTCCCATTGGATGGCACCTTAATGTTCAAAGTCGTTGCTTCAGCTGCGATGTACCTTATTATTTTAGTTCAGTTTGATAAAAGAAATTAA
Protein
MPLSLKQLELIYIKAFELKRDINKAFEAPILLTTMQCFHSIVSESHIIYHGAVMEPHMVLHSIMNCSVWILYQLFKLYILASTGHLLQEKIQHFSNLIHFHGKGLTVYGLFPLDGTLMFKVVASAAMYLIILVQFDKRN
Summary
Description
Gustatory receptor which mediates acceptance or avoidance behavior, depending on its substrates.
Dsx-dependent essential component of pheromone-driven courtship behavior (PubMed:12971900). Recognizes a female pheromone involved in the second step (tapping step) of the courtship display which is essential for efficient execution of the entire courtship sequence and timely mating (PubMed:12971900). Required for detection of the male sex pheromone CH503 which is transferred from males to females during mating and inhibits courtship behavior by other males (PubMed:26083710). Gr68a-expressing neurons in the male foreleg relay signals to the suboesophageal zone (SEZ) and courtship suppression is mediated by the release of the neuropeptide tachykinin from a cluster of 8-10 neurons in the SEZ (PubMed:26083710).
Dsx-dependent essential component of pheromone-driven courtship behavior (PubMed:12971900). Recognizes a female pheromone involved in the second step (tapping step) of the courtship display which is essential for efficient execution of the entire courtship sequence and timely mating (PubMed:12971900). Required for detection of the male sex pheromone CH503 which is transferred from males to females during mating and inhibits courtship behavior by other males (PubMed:26083710). Gr68a-expressing neurons in the male foreleg relay signals to the suboesophageal zone (SEZ) and courtship suppression is mediated by the release of the neuropeptide tachykinin from a cluster of 8-10 neurons in the SEZ (PubMed:26083710).
Similarity
Belongs to the insect chemoreceptor superfamily. Gustatory receptor (GR) family.
Belongs to the insect chemoreceptor superfamily. Gustatory receptor (GR) family. Gr21a subfamily.
Belongs to the insect chemoreceptor superfamily. Gustatory receptor (GR) family. Gr21a subfamily.
Keywords
Behavior
Cell membrane
Complete proteome
Glycoprotein
Membrane
Receptor
Reference proteome
Transducer
Transmembrane
Transmembrane helix
Feature
chain Gustatory receptor 68a
Uniprot
F8WPI3
A0A3S2PEY1
A0A0V0J185
A0A223HD94
A0A345BF03
A0A2H1W1K2
+ More
A0A0V0J2M4 A0A345BF07 A0A0V0J1G5 A0A1X9P7N0 A0A223HDH3 A0A223HDA0 A0A223HCX5 A0A0K8TVN5 A0A223HDI3 A0A0V0J1A1 A0A223HCW4 A0A223HDG2 A0A223HDA5 A0A223HDI0 A0A223HDA9 A0A0K8TUH2 A0A223HCW5 K7IUE9 A0A182UZ77 A0A1J1J099 B0WFG0 A0A3L9LWA8 A0A232FAC2 A0A1S4KKK6 A0A1S4JG72 A0A067RKK0 A0A1J1IXS9 J9HZ11 J9HSV8 J9EA72 A0A1S4FHY1 J9HSW2 Q170E5 A0A1S4FI97 J9HIL0 A0A1S4FHZ2 J9HFV3 J9HIL4 A0A1S4FI52 J9HZ06 A0A2P9JYB9 A0A1S4FHZ7 A0A182GFK6 A0A182QNT4 A0A182FRY3 A0A084W085 A5JHN2 A0A182YNA0 K7J4H3 A0A182XNX6 A0A154PGK2 B3NH01 A0A182HK22 A0A182VW97 A0A182NR18 A0A182WQQ3 A0A2R7WZH0 A5JHN3 A0A182R551 A2AXB7 A0A182FZN8 A0A067QUQ3 C9QPG7 Q9VTN0
A0A0V0J2M4 A0A345BF07 A0A0V0J1G5 A0A1X9P7N0 A0A223HDH3 A0A223HDA0 A0A223HCX5 A0A0K8TVN5 A0A223HDI3 A0A0V0J1A1 A0A223HCW4 A0A223HDG2 A0A223HDA5 A0A223HDI0 A0A223HDA9 A0A0K8TUH2 A0A223HCW5 K7IUE9 A0A182UZ77 A0A1J1J099 B0WFG0 A0A3L9LWA8 A0A232FAC2 A0A1S4KKK6 A0A1S4JG72 A0A067RKK0 A0A1J1IXS9 J9HZ11 J9HSV8 J9EA72 A0A1S4FHY1 J9HSW2 Q170E5 A0A1S4FI97 J9HIL0 A0A1S4FHZ2 J9HFV3 J9HIL4 A0A1S4FI52 J9HZ06 A0A2P9JYB9 A0A1S4FHZ7 A0A182GFK6 A0A182QNT4 A0A182FRY3 A0A084W085 A5JHN2 A0A182YNA0 K7J4H3 A0A182XNX6 A0A154PGK2 B3NH01 A0A182HK22 A0A182VW97 A0A182NR18 A0A182WQQ3 A0A2R7WZH0 A5JHN3 A0A182R551 A2AXB7 A0A182FZN8 A0A067QUQ3 C9QPG7 Q9VTN0
Pubmed
EMBL
AB600837
BAK52800.1
RSAL01000063
RVE49494.1
GDKB01000015
JAP38481.1
+ More
KY283690 AST36349.1 MG820656 AXF48827.1 ODYU01005767 SOQ46961.1 GDKB01000016 JAP38480.1 MG820660 AXF48831.1 GDKB01000018 JAP38478.1 KX084517 ARO76472.1 KY283770 AST36429.1 KY283693 AST36352.1 KY283561 AST36221.1 GCVX01000100 JAI18130.1 KY283772 AST36431.1 GDKB01000017 JAP38479.1 KY283554 KY283560 AST36214.1 AST36220.1 KY283771 AST36430.1 KY283692 AST36351.1 KY283769 AST36428.1 KY283694 AST36353.1 GCVX01000103 JAI18127.1 KY283559 AST36219.1 AAZX01005844 CVRI01000065 CRL05760.1 DS231917 EDS26197.1 KB465648 RLZ02140.1 NNAY01000540 OXU27796.1 KK852415 KDR24352.1 CVRI01000063 CRL04888.1 CH477475 EJY57725.1 EJY57718.1 EJY57721.1 EJY57723.1 EAT40327.2 EJY57719.1 EJY57722.1 EJY57724.1 EJY57720.1 KY817111 AVH87327.1 JXUM01059729 JXUM01059730 KQ562069 KXJ76746.1 AXCN02001482 ATLV01019096 KE525262 KFB43629.1 EF502095 ABQ57301.1 KQ434899 KZC10989.1 CH954178 EDV51458.1 APCN01000814 KK856124 PTY24943.1 EF502096 ABQ57302.1 AM292376 CAL23188.2 KK852927 KDR13678.1 BT100081 ACX61622.3 AE014296
KY283690 AST36349.1 MG820656 AXF48827.1 ODYU01005767 SOQ46961.1 GDKB01000016 JAP38480.1 MG820660 AXF48831.1 GDKB01000018 JAP38478.1 KX084517 ARO76472.1 KY283770 AST36429.1 KY283693 AST36352.1 KY283561 AST36221.1 GCVX01000100 JAI18130.1 KY283772 AST36431.1 GDKB01000017 JAP38479.1 KY283554 KY283560 AST36214.1 AST36220.1 KY283771 AST36430.1 KY283692 AST36351.1 KY283769 AST36428.1 KY283694 AST36353.1 GCVX01000103 JAI18127.1 KY283559 AST36219.1 AAZX01005844 CVRI01000065 CRL05760.1 DS231917 EDS26197.1 KB465648 RLZ02140.1 NNAY01000540 OXU27796.1 KK852415 KDR24352.1 CVRI01000063 CRL04888.1 CH477475 EJY57725.1 EJY57718.1 EJY57721.1 EJY57723.1 EAT40327.2 EJY57719.1 EJY57722.1 EJY57724.1 EJY57720.1 KY817111 AVH87327.1 JXUM01059729 JXUM01059730 KQ562069 KXJ76746.1 AXCN02001482 ATLV01019096 KE525262 KFB43629.1 EF502095 ABQ57301.1 KQ434899 KZC10989.1 CH954178 EDV51458.1 APCN01000814 KK856124 PTY24943.1 EF502096 ABQ57302.1 AM292376 CAL23188.2 KK852927 KDR13678.1 BT100081 ACX61622.3 AE014296
Proteomes
Pfam
PF08395 7tm_7
Interpro
IPR013604
7TM_chemorcpt
ProteinModelPortal
F8WPI3
A0A3S2PEY1
A0A0V0J185
A0A223HD94
A0A345BF03
A0A2H1W1K2
+ More
A0A0V0J2M4 A0A345BF07 A0A0V0J1G5 A0A1X9P7N0 A0A223HDH3 A0A223HDA0 A0A223HCX5 A0A0K8TVN5 A0A223HDI3 A0A0V0J1A1 A0A223HCW4 A0A223HDG2 A0A223HDA5 A0A223HDI0 A0A223HDA9 A0A0K8TUH2 A0A223HCW5 K7IUE9 A0A182UZ77 A0A1J1J099 B0WFG0 A0A3L9LWA8 A0A232FAC2 A0A1S4KKK6 A0A1S4JG72 A0A067RKK0 A0A1J1IXS9 J9HZ11 J9HSV8 J9EA72 A0A1S4FHY1 J9HSW2 Q170E5 A0A1S4FI97 J9HIL0 A0A1S4FHZ2 J9HFV3 J9HIL4 A0A1S4FI52 J9HZ06 A0A2P9JYB9 A0A1S4FHZ7 A0A182GFK6 A0A182QNT4 A0A182FRY3 A0A084W085 A5JHN2 A0A182YNA0 K7J4H3 A0A182XNX6 A0A154PGK2 B3NH01 A0A182HK22 A0A182VW97 A0A182NR18 A0A182WQQ3 A0A2R7WZH0 A5JHN3 A0A182R551 A2AXB7 A0A182FZN8 A0A067QUQ3 C9QPG7 Q9VTN0
A0A0V0J2M4 A0A345BF07 A0A0V0J1G5 A0A1X9P7N0 A0A223HDH3 A0A223HDA0 A0A223HCX5 A0A0K8TVN5 A0A223HDI3 A0A0V0J1A1 A0A223HCW4 A0A223HDG2 A0A223HDA5 A0A223HDI0 A0A223HDA9 A0A0K8TUH2 A0A223HCW5 K7IUE9 A0A182UZ77 A0A1J1J099 B0WFG0 A0A3L9LWA8 A0A232FAC2 A0A1S4KKK6 A0A1S4JG72 A0A067RKK0 A0A1J1IXS9 J9HZ11 J9HSV8 J9EA72 A0A1S4FHY1 J9HSW2 Q170E5 A0A1S4FI97 J9HIL0 A0A1S4FHZ2 J9HFV3 J9HIL4 A0A1S4FI52 J9HZ06 A0A2P9JYB9 A0A1S4FHZ7 A0A182GFK6 A0A182QNT4 A0A182FRY3 A0A084W085 A5JHN2 A0A182YNA0 K7J4H3 A0A182XNX6 A0A154PGK2 B3NH01 A0A182HK22 A0A182VW97 A0A182NR18 A0A182WQQ3 A0A2R7WZH0 A5JHN3 A0A182R551 A2AXB7 A0A182FZN8 A0A067QUQ3 C9QPG7 Q9VTN0
Ontologies
GO
PANTHER
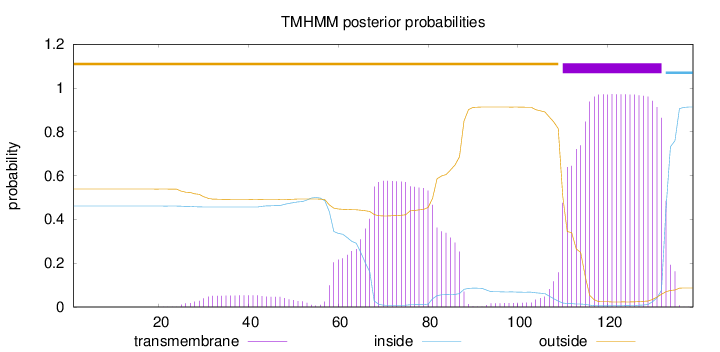
Topology
Subcellular location
Cell membrane
Length:
139
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
35.38215
Exp number, first 60 AAs:
1.74558
Total prob of N-in:
0.46142
outside
1 - 109
TMhelix
110 - 132
inside
133 - 139
Population Genetic Test Statistics
Pi
287.318492
Theta
219.361733
Tajima's D
0.975464
CLR
0
CSRT
0.655117244137793
Interpretation
Uncertain
