Gene
KWMTBOMO04409
Pre Gene Modal
BGIBMGA005402
Annotation
PREDICTED:_probable_ATP-dependent_DNA_helicase_HFM1_[Amyelois_transitella]
Full name
Probable ATP-dependent DNA helicase HFM1
Location in the cell
PlasmaMembrane Reliability : 1.979
Sequence
CDS
ATGTTTCCGCACTTGCAGTCTCTAATCATTAGCGGCGTTGGCTGTCACCACGCTGGGCTCCTATTCGAAGAGAGAATGTACATTGAGAGCGCGTTCAGAAACAGAGATCTACCAATACTTATTACCACCACCACGCTCGCTATGGGCGTAAATTTACCAGCTCATTTAGTGATCATCAAGAATACACAGCAATACGTTAACGGAGCTTATCAGGAATACAGTATAAGTACGGTATTGCAAATGGTCGGTCGAGCTGGACGACCGCAATTCGACACCGAAGCTACCGCCGTCATAATGACGAGACTTGCCGATAAGGTGAGTCCTGATATTAATTAG
Protein
MFPHLQSLIISGVGCHHAGLLFEERMYIESAFRNRDLPILITTTTLAMGVNLPAHLVIIKNTQQYVNGAYQEYSISTVLQMVGRAGRPQFDTEATAVIMTRLADKVSPDIN
Summary
Description
Required for crossover formation and complete synapsis of homologous chromosomes during meiosis.
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Similarity
Belongs to the helicase family. SKI2 subfamily.
Keywords
ATP-binding
Complete proteome
Helicase
Hydrolase
Meiosis
Nucleotide-binding
Reference proteome
Feature
chain Probable ATP-dependent DNA helicase HFM1
Uniprot
H9J7B0
A0A2H1W4Q4
A0A2A4IWG5
A0A194PWH8
A0A212EHY6
A0A3S2NV24
+ More
W4XYW1 A0A2R7VNT0 G3WP41 W4XIW2 K7EBL3 A0A383ZMP0 A0A094LLJ3 A0A0Q3Q1S2 A0A3Q0GH13 A0A3Q0GDK2 G1KUQ1 H0VA28 M3XZQ8 H3CNC9 F7ARR8 A0A286ZUN5 F1LWY1 F1S4E9 G5ASI6 A0A287BMJ1 A0A093HRD6 A0A151M532 A0A091MH88 A0A151M534 A0A091KPU4 A0A383ZNB6 A0A1L8GMZ3 A0A2U3ZHX7 F6WRW2 A0A340Y4G3 A0A384C3M7 A0A2U3ZHX9 G1L0V4 A0A2I0MDY0 G3PSW6 A0A2I0UMQ7 A0A3Q7QJB0 A0A2U3ZHY8 A0A3Q7VWX0 A0A1S3HGY3 A0A2Y9SAA3 E2R5U9 D2HUC3 A0A1V4JYQ1 E2RA34 A0A091QWS2 A0A2Y9LHR9 A0A1V4JYN5 A0A1V4JYQ2 A0A1V4K0C3 A0A2Y9ITI2 A0A3Q7S1S8 A0A341BCJ9 G3I0Y7 A0A3L7H9E0 A0A2Y9LPT0 A0A091XSM1 A0A2Y9LJZ4 A0A2U4A133 A0A091HRT4 A0A091FKX5 A0A091TN48 A0A2Y9LHG4 A0A091QL31 A0A1S2ZUG5 A0A091CP49 A0A1U7UKT7 W5MC30 A0A2U3XBS1 A0A3Q0D1F8 A2RUV5 A0A091NMI4 A0A2Y9H2I8 A0A2U3XBP4 F1MFR1 A0A093IT05 A0A091T3U2 A0A2U9BJU0 A0A3Q1IZJ9 H0Z6Z6 A0A3Q1JBL2 A0A091MBM7 A0A0A0AJ85 A0A087QHI4 A0A3L8SPX5 H0XZQ2 U3JGG5 A0A091L989 A0A2Y9RCB7 A0A3Q1J4S4 A0A093QF48 A0A3P8T6N5 A0A2D0SW76 A0A2D0SVQ9
W4XYW1 A0A2R7VNT0 G3WP41 W4XIW2 K7EBL3 A0A383ZMP0 A0A094LLJ3 A0A0Q3Q1S2 A0A3Q0GH13 A0A3Q0GDK2 G1KUQ1 H0VA28 M3XZQ8 H3CNC9 F7ARR8 A0A286ZUN5 F1LWY1 F1S4E9 G5ASI6 A0A287BMJ1 A0A093HRD6 A0A151M532 A0A091MH88 A0A151M534 A0A091KPU4 A0A383ZNB6 A0A1L8GMZ3 A0A2U3ZHX7 F6WRW2 A0A340Y4G3 A0A384C3M7 A0A2U3ZHX9 G1L0V4 A0A2I0MDY0 G3PSW6 A0A2I0UMQ7 A0A3Q7QJB0 A0A2U3ZHY8 A0A3Q7VWX0 A0A1S3HGY3 A0A2Y9SAA3 E2R5U9 D2HUC3 A0A1V4JYQ1 E2RA34 A0A091QWS2 A0A2Y9LHR9 A0A1V4JYN5 A0A1V4JYQ2 A0A1V4K0C3 A0A2Y9ITI2 A0A3Q7S1S8 A0A341BCJ9 G3I0Y7 A0A3L7H9E0 A0A2Y9LPT0 A0A091XSM1 A0A2Y9LJZ4 A0A2U4A133 A0A091HRT4 A0A091FKX5 A0A091TN48 A0A2Y9LHG4 A0A091QL31 A0A1S2ZUG5 A0A091CP49 A0A1U7UKT7 W5MC30 A0A2U3XBS1 A0A3Q0D1F8 A2RUV5 A0A091NMI4 A0A2Y9H2I8 A0A2U3XBP4 F1MFR1 A0A093IT05 A0A091T3U2 A0A2U9BJU0 A0A3Q1IZJ9 H0Z6Z6 A0A3Q1JBL2 A0A091MBM7 A0A0A0AJ85 A0A087QHI4 A0A3L8SPX5 H0XZQ2 U3JGG5 A0A091L989 A0A2Y9RCB7 A0A3Q1J4S4 A0A093QF48 A0A3P8T6N5 A0A2D0SW76 A0A2D0SVQ9
EC Number
3.6.4.12
Pubmed
EMBL
BABH01013596
ODYU01006288
SOQ48003.1
NWSH01005710
PCG64019.1
KQ459589
+ More
KPI97677.1 AGBW02014758 OWR41095.1 RSAL01000063 RVE49498.1 AAGJ04028426 KK854008 PTY09197.1 AEFK01152901 AEFK01152902 AEFK01152903 AEFK01152904 KL359789 KFZ64664.1 LMAW01002002 KQK82116.1 AAKN02004044 AAKN02004045 AAKN02004046 AAKN02004047 AEYP01031963 AEYP01031964 AEMK02000025 AABR07014161 JH166735 EHA99996.1 KL206716 KFV85228.1 AKHW03006584 KYO19627.1 KK828177 KFP74399.1 KYO19626.1 KK537306 KFP29804.1 CM004472 OCT85181.1 ACTA01066455 ACTA01074455 ACTA01082455 ACTA01090455 ACTA01098455 ACTA01106455 ACTA01114455 ACTA01122455 AKCR02000018 PKK27866.1 KZ505681 PKU47327.1 AAEX03004823 GL193394 EFB13360.1 LSYS01005497 OPJ77339.1 KK707602 KFQ31339.1 OPJ77340.1 OPJ77338.1 OPJ77337.1 JH001037 EGW07141.1 RAZU01000247 RLQ62559.1 KK735808 KFR15916.1 KL217788 KFO98646.1 KL447073 KFO69596.1 KK479827 KFQ60141.1 KK800274 KFQ28265.1 KN125242 KFO18955.1 AHAT01018350 BC133060 BC121498 KL387128 KFP90307.1 KK566757 KFW04798.1 KK437011 KFQ69021.1 CP026249 AWP04338.1 ABQF01035428 ABQF01035429 ABQF01035430 KK528167 KFP68886.1 KL871737 KGL93095.1 KL225597 KFM00688.1 QUSF01000010 RLW05663.1 AAQR03023018 AAQR03023019 AAQR03023020 AAQR03023021 AAQR03023022 AAQR03023023 AGTO01001511 KL307548 KFP52432.1 KL416641 KFW82722.1
KPI97677.1 AGBW02014758 OWR41095.1 RSAL01000063 RVE49498.1 AAGJ04028426 KK854008 PTY09197.1 AEFK01152901 AEFK01152902 AEFK01152903 AEFK01152904 KL359789 KFZ64664.1 LMAW01002002 KQK82116.1 AAKN02004044 AAKN02004045 AAKN02004046 AAKN02004047 AEYP01031963 AEYP01031964 AEMK02000025 AABR07014161 JH166735 EHA99996.1 KL206716 KFV85228.1 AKHW03006584 KYO19627.1 KK828177 KFP74399.1 KYO19626.1 KK537306 KFP29804.1 CM004472 OCT85181.1 ACTA01066455 ACTA01074455 ACTA01082455 ACTA01090455 ACTA01098455 ACTA01106455 ACTA01114455 ACTA01122455 AKCR02000018 PKK27866.1 KZ505681 PKU47327.1 AAEX03004823 GL193394 EFB13360.1 LSYS01005497 OPJ77339.1 KK707602 KFQ31339.1 OPJ77340.1 OPJ77338.1 OPJ77337.1 JH001037 EGW07141.1 RAZU01000247 RLQ62559.1 KK735808 KFR15916.1 KL217788 KFO98646.1 KL447073 KFO69596.1 KK479827 KFQ60141.1 KK800274 KFQ28265.1 KN125242 KFO18955.1 AHAT01018350 BC133060 BC121498 KL387128 KFP90307.1 KK566757 KFW04798.1 KK437011 KFQ69021.1 CP026249 AWP04338.1 ABQF01035428 ABQF01035429 ABQF01035430 KK528167 KFP68886.1 KL871737 KGL93095.1 KL225597 KFM00688.1 QUSF01000010 RLW05663.1 AAQR03023018 AAQR03023019 AAQR03023020 AAQR03023021 AAQR03023022 AAQR03023023 AGTO01001511 KL307548 KFP52432.1 KL416641 KFW82722.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000283053
UP000007110
+ More
UP000007648 UP000002279 UP000261681 UP000051836 UP000189705 UP000001646 UP000005447 UP000000715 UP000007303 UP000002280 UP000008227 UP000002494 UP000006813 UP000053584 UP000050525 UP000186698 UP000245340 UP000002281 UP000265300 UP000261680 UP000008912 UP000053872 UP000007635 UP000286641 UP000286642 UP000085678 UP000002254 UP000190648 UP000248483 UP000248482 UP000286640 UP000252040 UP000001075 UP000273346 UP000053605 UP000245320 UP000054308 UP000053760 UP000079721 UP000028990 UP000189704 UP000018468 UP000245341 UP000189706 UP000008143 UP000248481 UP000009136 UP000246464 UP000265040 UP000007754 UP000053858 UP000053286 UP000276834 UP000005225 UP000016665 UP000248480 UP000265080 UP000221080
UP000007648 UP000002279 UP000261681 UP000051836 UP000189705 UP000001646 UP000005447 UP000000715 UP000007303 UP000002280 UP000008227 UP000002494 UP000006813 UP000053584 UP000050525 UP000186698 UP000245340 UP000002281 UP000265300 UP000261680 UP000008912 UP000053872 UP000007635 UP000286641 UP000286642 UP000085678 UP000002254 UP000190648 UP000248483 UP000248482 UP000286640 UP000252040 UP000001075 UP000273346 UP000053605 UP000245320 UP000054308 UP000053760 UP000079721 UP000028990 UP000189704 UP000018468 UP000245341 UP000189706 UP000008143 UP000248481 UP000009136 UP000246464 UP000265040 UP000007754 UP000053858 UP000053286 UP000276834 UP000005225 UP000016665 UP000248480 UP000265080 UP000221080
Interpro
CDD
ProteinModelPortal
H9J7B0
A0A2H1W4Q4
A0A2A4IWG5
A0A194PWH8
A0A212EHY6
A0A3S2NV24
+ More
W4XYW1 A0A2R7VNT0 G3WP41 W4XIW2 K7EBL3 A0A383ZMP0 A0A094LLJ3 A0A0Q3Q1S2 A0A3Q0GH13 A0A3Q0GDK2 G1KUQ1 H0VA28 M3XZQ8 H3CNC9 F7ARR8 A0A286ZUN5 F1LWY1 F1S4E9 G5ASI6 A0A287BMJ1 A0A093HRD6 A0A151M532 A0A091MH88 A0A151M534 A0A091KPU4 A0A383ZNB6 A0A1L8GMZ3 A0A2U3ZHX7 F6WRW2 A0A340Y4G3 A0A384C3M7 A0A2U3ZHX9 G1L0V4 A0A2I0MDY0 G3PSW6 A0A2I0UMQ7 A0A3Q7QJB0 A0A2U3ZHY8 A0A3Q7VWX0 A0A1S3HGY3 A0A2Y9SAA3 E2R5U9 D2HUC3 A0A1V4JYQ1 E2RA34 A0A091QWS2 A0A2Y9LHR9 A0A1V4JYN5 A0A1V4JYQ2 A0A1V4K0C3 A0A2Y9ITI2 A0A3Q7S1S8 A0A341BCJ9 G3I0Y7 A0A3L7H9E0 A0A2Y9LPT0 A0A091XSM1 A0A2Y9LJZ4 A0A2U4A133 A0A091HRT4 A0A091FKX5 A0A091TN48 A0A2Y9LHG4 A0A091QL31 A0A1S2ZUG5 A0A091CP49 A0A1U7UKT7 W5MC30 A0A2U3XBS1 A0A3Q0D1F8 A2RUV5 A0A091NMI4 A0A2Y9H2I8 A0A2U3XBP4 F1MFR1 A0A093IT05 A0A091T3U2 A0A2U9BJU0 A0A3Q1IZJ9 H0Z6Z6 A0A3Q1JBL2 A0A091MBM7 A0A0A0AJ85 A0A087QHI4 A0A3L8SPX5 H0XZQ2 U3JGG5 A0A091L989 A0A2Y9RCB7 A0A3Q1J4S4 A0A093QF48 A0A3P8T6N5 A0A2D0SW76 A0A2D0SVQ9
W4XYW1 A0A2R7VNT0 G3WP41 W4XIW2 K7EBL3 A0A383ZMP0 A0A094LLJ3 A0A0Q3Q1S2 A0A3Q0GH13 A0A3Q0GDK2 G1KUQ1 H0VA28 M3XZQ8 H3CNC9 F7ARR8 A0A286ZUN5 F1LWY1 F1S4E9 G5ASI6 A0A287BMJ1 A0A093HRD6 A0A151M532 A0A091MH88 A0A151M534 A0A091KPU4 A0A383ZNB6 A0A1L8GMZ3 A0A2U3ZHX7 F6WRW2 A0A340Y4G3 A0A384C3M7 A0A2U3ZHX9 G1L0V4 A0A2I0MDY0 G3PSW6 A0A2I0UMQ7 A0A3Q7QJB0 A0A2U3ZHY8 A0A3Q7VWX0 A0A1S3HGY3 A0A2Y9SAA3 E2R5U9 D2HUC3 A0A1V4JYQ1 E2RA34 A0A091QWS2 A0A2Y9LHR9 A0A1V4JYN5 A0A1V4JYQ2 A0A1V4K0C3 A0A2Y9ITI2 A0A3Q7S1S8 A0A341BCJ9 G3I0Y7 A0A3L7H9E0 A0A2Y9LPT0 A0A091XSM1 A0A2Y9LJZ4 A0A2U4A133 A0A091HRT4 A0A091FKX5 A0A091TN48 A0A2Y9LHG4 A0A091QL31 A0A1S2ZUG5 A0A091CP49 A0A1U7UKT7 W5MC30 A0A2U3XBS1 A0A3Q0D1F8 A2RUV5 A0A091NMI4 A0A2Y9H2I8 A0A2U3XBP4 F1MFR1 A0A093IT05 A0A091T3U2 A0A2U9BJU0 A0A3Q1IZJ9 H0Z6Z6 A0A3Q1JBL2 A0A091MBM7 A0A0A0AJ85 A0A087QHI4 A0A3L8SPX5 H0XZQ2 U3JGG5 A0A091L989 A0A2Y9RCB7 A0A3Q1J4S4 A0A093QF48 A0A3P8T6N5 A0A2D0SW76 A0A2D0SVQ9
PDB
5ZWO
E-value=6.59077e-21,
Score=241
Ontologies
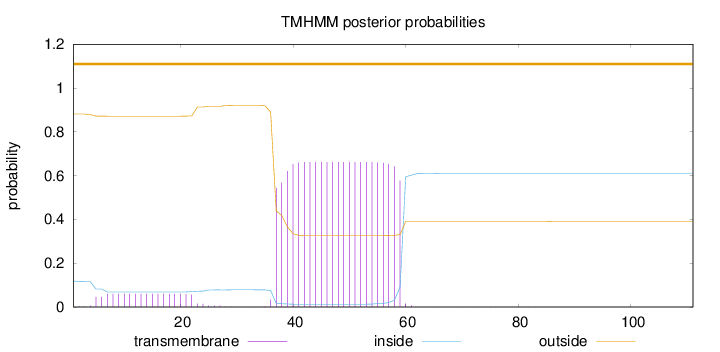
Topology
Length:
111
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
16.02414
Exp number, first 60 AAs:
16.00001
Total prob of N-in:
0.11873
POSSIBLE N-term signal
sequence
outside
1 - 111
Population Genetic Test Statistics
Pi
460.110845
Theta
197.15765
Tajima's D
3.535916
CLR
0.950525
CSRT
0.994750262486876
Interpretation
Uncertain
