Gene
KWMTBOMO04408
Annotation
PREDICTED:_uncharacterized_protein_LOC101741884_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.756 Nuclear Reliability : 1.712
Sequence
CDS
ATGGGTGCTGCCTGCTATGAAGTCTGGAAAACCATAGTGAAGAAAGTGGGGGGTTCTCCAAATTGTTGGAGAGATGTTGGCCATTTATTAGGTGTTACACAGGATGACCTAAATTACATAATGAATTCTTTAAAAGATGACCCAGTGGATATGGTATTGAAAGTATTCAGACAAAATGAAAAAGCTACTATAAATAAAATAGTTGATGCATTCATAAAACTGCAGAGGTATGACATCTTGTTAGCAATTGAAAATCCACTGTGTACTATGGCTGAATATTTTAATAAAGATGATAGTGGTTATCAAAGTGATGGTAAGATAACTGGTAAAAAGGAAATTGTTACACTCAAAAACCTACCCAATGATCTACCTGCAGCTTTAAATAAAAATATAGTACTTCAAAACAATGACCCTAAAAGACCTAAAGAAACGTTACAGCCCCCAGTTAGTAAACAGGAGCCAGCAGAAAAAGAAAGCCCTATTTTATTCTTAACTTATGCTCAAGATGGTCTACCAACTGCACTTAACATACAGGAGTATGTTAGTAATTGGACGGAAATACCAAATGTTAAAGTCATAACATTAAACAATAGAAGAGAAGAAACTTACCAGAATCCTGAAAAATTTATAAGAGAATATTTTGAAAAGGCTGACTACATTGTGCCAATCATAACAACAGGATATATAAATGAAATAACCTCAAACAACAAAAATATACCAAATACAACAGAGAATTTGGATCACAAATATGTTAATTTCATATATAATCTCATTGTTAACAATTATATTCATGCAACTGGTTGTCTTAACAGAAAAGTAAGAAGTGTGCTGCCACAGAATGCAAATGTTGATCTATTTTCACGTATGACAATGTATCCTGATTTATTGCCTTGGACATACGAGACAAACTTTGATGAACAGTTTCAAGAGTTTTTGAGAAAAGAACCTTGA
Protein
MGAACYEVWKTIVKKVGGSPNCWRDVGHLLGVTQDDLNYIMNSLKDDPVDMVLKVFRQNEKATINKIVDAFIKLQRYDILLAIENPLCTMAEYFNKDDSGYQSDGKITGKKEIVTLKNLPNDLPAALNKNIVLQNNDPKRPKETLQPPVSKQEPAEKESPILFLTYAQDGLPTALNIQEYVSNWTEIPNVKVITLNNRREETYQNPEKFIREYFEKADYIVPIITTGYINEITSNNKNIPNTTENLDHKYVNFIYNLIVNNYIHATGCLNRKVRSVLPQNANVDLFSRMTMYPDLLPWTYETNFDEQFQEFLRKEP
Summary
Uniprot
A0A2H1W674
A0A2A4IV09
I3RJG4
A0A2A4IWZ8
A0A0L7L0L1
A0A212EHZ0
+ More
A0A1E1WLR0 A0A0N1IJX3 A0A194PWC2 A0A2A4IU57 S4PV42 A0A0C9PVA5 A0A195F5G5 A0A026X505 A0A151J603 A0A195D1H1 A0A151X5G0 A0A3L8DNJ0 A0A067RP46 A0A195AZU3 A0A0V0G369 A0A158NH50 A0A0P4VMZ0 T1HTY5 E9IMW3 A0A069DYU1 K7JJQ8 A0A2A3ESZ6 A0A170XN98 A0A023EXR8 A0A171B9S8 A0A088AF34 A0A2J7PED6 F8UXA2 A0A154P1X5 A0A224XRX0 A0A0L7QYK7 A0A1S3CZX2 A0A1B6C388 A0A146MEB9 A0A146L0V1 A0A0K8SA56 A0A1B6DW91 A0A0M8ZWG7 A0A232FNE2 A0A0P5VJ13 A0A0P6EGS3 A0A0P6B1Z3 E9GDJ6 A0A0P5FA45 A0A0N8BXZ5 A0A1B6MJR1 A0A0P5YJ44 A0A1B6H507 A0A1B6HZE6
A0A1E1WLR0 A0A0N1IJX3 A0A194PWC2 A0A2A4IU57 S4PV42 A0A0C9PVA5 A0A195F5G5 A0A026X505 A0A151J603 A0A195D1H1 A0A151X5G0 A0A3L8DNJ0 A0A067RP46 A0A195AZU3 A0A0V0G369 A0A158NH50 A0A0P4VMZ0 T1HTY5 E9IMW3 A0A069DYU1 K7JJQ8 A0A2A3ESZ6 A0A170XN98 A0A023EXR8 A0A171B9S8 A0A088AF34 A0A2J7PED6 F8UXA2 A0A154P1X5 A0A224XRX0 A0A0L7QYK7 A0A1S3CZX2 A0A1B6C388 A0A146MEB9 A0A146L0V1 A0A0K8SA56 A0A1B6DW91 A0A0M8ZWG7 A0A232FNE2 A0A0P5VJ13 A0A0P6EGS3 A0A0P6B1Z3 E9GDJ6 A0A0P5FA45 A0A0N8BXZ5 A0A1B6MJR1 A0A0P5YJ44 A0A1B6H507 A0A1B6HZE6
Pubmed
EMBL
ODYU01006288
SOQ48004.1
NWSH01005955
PCG63807.1
JQ687409
AFK24444.1
+ More
PCG63804.1 JTDY01003776 KOB69022.1 AGBW02014758 OWR41094.1 GDQN01003263 JAT87791.1 KQ459761 KPJ20132.1 KQ459589 KPI97676.1 NWSH01008053 PCG62712.1 GAIX01009193 JAA83367.1 GBYB01005343 GBYB01009719 GBYB01009722 JAG75110.1 JAG79486.1 JAG79489.1 KQ981768 KYN35840.1 KK107020 EZA62509.1 KQ979900 KYN18653.1 KQ976973 KYN06717.1 KQ982510 KYQ55636.1 QOIP01000006 RLU21378.1 KK852508 KDR22395.1 KQ976698 KYM77479.1 GECL01003585 JAP02539.1 ADTU01015577 GDKW01000036 JAI56559.1 ACPB03011671 GL764255 EFZ18078.1 GBGD01001800 JAC87089.1 KZ288185 PBC34903.1 GEMB01004232 JAR99037.1 GBBI01004931 JAC13781.1 GEMB01000096 JAS03015.1 NEVH01026101 PNF14700.1 JF965014 AEJ08192.1 KQ434803 KZC05935.1 GFTR01005697 JAW10729.1 KQ414688 KOC63641.1 GEDC01029549 JAS07749.1 GDHC01001719 JAQ16910.1 GDHC01017747 JAQ00882.1 GBRD01015650 JAG50176.1 GEDC01007344 JAS29954.1 KQ435821 KOX72337.1 NNAY01000011 OXU32039.1 GDIP01099306 LRGB01002672 JAM04409.1 KZS06601.1 GDIQ01062817 JAN31920.1 GDIP01021177 JAM82538.1 GL732540 EFX82465.1 GDIQ01257618 JAJ94106.1 GDIQ01133899 JAL17827.1 GEBQ01003770 JAT36207.1 GDIP01057102 JAM46613.1 GECZ01000016 JAS69753.1 GECU01027660 JAS80046.1
PCG63804.1 JTDY01003776 KOB69022.1 AGBW02014758 OWR41094.1 GDQN01003263 JAT87791.1 KQ459761 KPJ20132.1 KQ459589 KPI97676.1 NWSH01008053 PCG62712.1 GAIX01009193 JAA83367.1 GBYB01005343 GBYB01009719 GBYB01009722 JAG75110.1 JAG79486.1 JAG79489.1 KQ981768 KYN35840.1 KK107020 EZA62509.1 KQ979900 KYN18653.1 KQ976973 KYN06717.1 KQ982510 KYQ55636.1 QOIP01000006 RLU21378.1 KK852508 KDR22395.1 KQ976698 KYM77479.1 GECL01003585 JAP02539.1 ADTU01015577 GDKW01000036 JAI56559.1 ACPB03011671 GL764255 EFZ18078.1 GBGD01001800 JAC87089.1 KZ288185 PBC34903.1 GEMB01004232 JAR99037.1 GBBI01004931 JAC13781.1 GEMB01000096 JAS03015.1 NEVH01026101 PNF14700.1 JF965014 AEJ08192.1 KQ434803 KZC05935.1 GFTR01005697 JAW10729.1 KQ414688 KOC63641.1 GEDC01029549 JAS07749.1 GDHC01001719 JAQ16910.1 GDHC01017747 JAQ00882.1 GBRD01015650 JAG50176.1 GEDC01007344 JAS29954.1 KQ435821 KOX72337.1 NNAY01000011 OXU32039.1 GDIP01099306 LRGB01002672 JAM04409.1 KZS06601.1 GDIQ01062817 JAN31920.1 GDIP01021177 JAM82538.1 GL732540 EFX82465.1 GDIQ01257618 JAJ94106.1 GDIQ01133899 JAL17827.1 GEBQ01003770 JAT36207.1 GDIP01057102 JAM46613.1 GECZ01000016 JAS69753.1 GECU01027660 JAS80046.1
Proteomes
UP000218220
UP000037510
UP000007151
UP000053240
UP000053268
UP000078541
+ More
UP000053097 UP000078492 UP000078542 UP000075809 UP000279307 UP000027135 UP000078540 UP000005205 UP000015103 UP000002358 UP000242457 UP000005203 UP000235965 UP000076502 UP000053825 UP000079169 UP000053105 UP000215335 UP000076858 UP000000305
UP000053097 UP000078492 UP000078542 UP000075809 UP000279307 UP000027135 UP000078540 UP000005205 UP000015103 UP000002358 UP000242457 UP000005203 UP000235965 UP000076502 UP000053825 UP000079169 UP000053105 UP000215335 UP000076858 UP000000305
Interpro
SUPFAM
SSF47986
SSF47986
Gene 3D
ProteinModelPortal
A0A2H1W674
A0A2A4IV09
I3RJG4
A0A2A4IWZ8
A0A0L7L0L1
A0A212EHZ0
+ More
A0A1E1WLR0 A0A0N1IJX3 A0A194PWC2 A0A2A4IU57 S4PV42 A0A0C9PVA5 A0A195F5G5 A0A026X505 A0A151J603 A0A195D1H1 A0A151X5G0 A0A3L8DNJ0 A0A067RP46 A0A195AZU3 A0A0V0G369 A0A158NH50 A0A0P4VMZ0 T1HTY5 E9IMW3 A0A069DYU1 K7JJQ8 A0A2A3ESZ6 A0A170XN98 A0A023EXR8 A0A171B9S8 A0A088AF34 A0A2J7PED6 F8UXA2 A0A154P1X5 A0A224XRX0 A0A0L7QYK7 A0A1S3CZX2 A0A1B6C388 A0A146MEB9 A0A146L0V1 A0A0K8SA56 A0A1B6DW91 A0A0M8ZWG7 A0A232FNE2 A0A0P5VJ13 A0A0P6EGS3 A0A0P6B1Z3 E9GDJ6 A0A0P5FA45 A0A0N8BXZ5 A0A1B6MJR1 A0A0P5YJ44 A0A1B6H507 A0A1B6HZE6
A0A1E1WLR0 A0A0N1IJX3 A0A194PWC2 A0A2A4IU57 S4PV42 A0A0C9PVA5 A0A195F5G5 A0A026X505 A0A151J603 A0A195D1H1 A0A151X5G0 A0A3L8DNJ0 A0A067RP46 A0A195AZU3 A0A0V0G369 A0A158NH50 A0A0P4VMZ0 T1HTY5 E9IMW3 A0A069DYU1 K7JJQ8 A0A2A3ESZ6 A0A170XN98 A0A023EXR8 A0A171B9S8 A0A088AF34 A0A2J7PED6 F8UXA2 A0A154P1X5 A0A224XRX0 A0A0L7QYK7 A0A1S3CZX2 A0A1B6C388 A0A146MEB9 A0A146L0V1 A0A0K8SA56 A0A1B6DW91 A0A0M8ZWG7 A0A232FNE2 A0A0P5VJ13 A0A0P6EGS3 A0A0P6B1Z3 E9GDJ6 A0A0P5FA45 A0A0N8BXZ5 A0A1B6MJR1 A0A0P5YJ44 A0A1B6H507 A0A1B6HZE6
Ontologies
GO
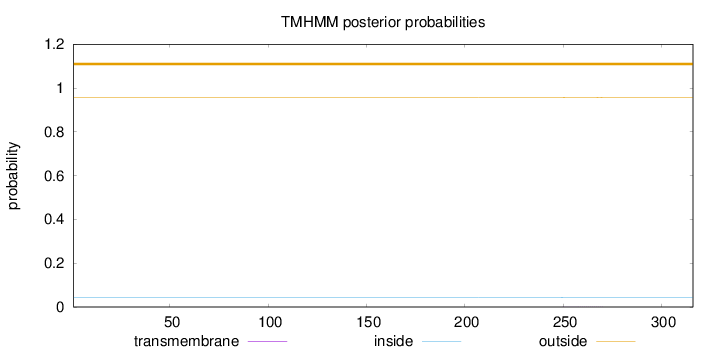
Topology
Length:
316
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01574
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04266
outside
1 - 316
Population Genetic Test Statistics
Pi
53.396864
Theta
156.386847
Tajima's D
-0.746254
CLR
104.215181
CSRT
0.189940502974851
Interpretation
Uncertain
