Gene
KWMTBOMO04406 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005398
Annotation
PREDICTED:_inositol_polyphosphate_5-phosphatase_K-like_isoform_X3_[Plutella_xylostella]
Location in the cell
Extracellular Reliability : 1.275
Sequence
CDS
ATGGATACTTTAAGATTCTATTTTGTAACCTACAATGTAGCTACAAAGGCACCAACACAGGATCTCAATGCTCTACTAGACTTTCCATCTCAGTACAACAAGAATAAGCCTTTGCCAGATTTTTATGTTATTGGGTTACAAGAAGTAAAATCACAGCCACAGAACATGCTAATGGACTCCCTATTCAGTGATCCGTGGACATCCACAGTCAACAAGATATTGTGCCGACAGGGATATATAATAGCCAAGAATACGAGATTGCAAGGTCTCTTGCTCTTGGTGTACACACAAATGAAACATGTTACACATTTAAGGGACATTGAAGCACAGTACACAAAAACAGGATTAGGTGGTATGTGGGGTAACAAAGGAGCGGTCAGCATAAGGTTCAATATATATGGCTGCTCCGTATGCTTAGTGAATTGTCATCTAACTGCGCACGAACATCTGTTGGCGGATCGGATTAATGACTACAACACTATAATTAAGCATCATCTGTACCATGTTAACGAGACCACAAACATATTGTATCATGATTATGTTTTCTGGATCGGCGACTTGAATTTTCGAACGGATCATCCGACCGGCAGCAGCCCTACGTCGGAAGAGATCGTCGCAAAATTGCAGAAGGTTGAGAAGGATAAATACGCTGCCTTGTTAAAACACGATCAGCTTTTAGCAGTCATGGATGCGGGGGAAGCTTTCTCAGAATTTTCCGAAGCCGATATAAAATTTCCCCCCACTTACAAATTCGTGATCGGCGGCGACGACTATGACCTCAAACGAAAACCATCATGGACAGACCGAGTCCTATACAAGGTAGTAGCGAATAACTATGAGAATGTAACGTTAAGGGCGGATCTTGTTTCATATAATCACATGCCGCACTACACACTGAGTGACCACAAGCCAGTTGTGGCGCAATTCAATATTAAGGCGTTTGCAAATTACACTGAGAAGACAGTCGAGTTCGCTCCGATAAATCGAGTTTGGTATATAGGAGACGCCGACTTCAGGACCCAGTGTACCCTCACGCCCGATATCGAAGTTAATCCGGGCGATTGGATCGGTATATATGATGCAAACTTCCACAACCTAGACGACTACATTTCATACGAATATTTAGCTAAGGTCGGTCTGTCTGTACGGCCGGCTAGTGACGGACAGCCGAGGACGATCACCCTGAGCTTTCCCGTGGGCAGCGGCGTGAGGACGCCGGGCTTCTACAGGTTCTTGTATTTCAGCCAGCCCAACAGCGACGTGCGGAGTGTCTTAGGTATATCCGAGCCCTTCGAAGTGAGCAGCAAAGAGGACAAGCTGGTCACTCTGGGCACAAGCGAAGAGCCGTCGACTAGTTCCTCGCCCGGCCAGCCCAAGTCGGCGACCGCCAGCACCGACATCTTCAACGACTTCACCGGTCTGGACGTGGCCAAGTTGTCCAGGCATTTCTCGAACGATCTCAGCATCGACTGA
Protein
MDTLRFYFVTYNVATKAPTQDLNALLDFPSQYNKNKPLPDFYVIGLQEVKSQPQNMLMDSLFSDPWTSTVNKILCRQGYIIAKNTRLQGLLLLVYTQMKHVTHLRDIEAQYTKTGLGGMWGNKGAVSIRFNIYGCSVCLVNCHLTAHEHLLADRINDYNTIIKHHLYHVNETTNILYHDYVFWIGDLNFRTDHPTGSSPTSEEIVAKLQKVEKDKYAALLKHDQLLAVMDAGEAFSEFSEADIKFPPTYKFVIGGDDYDLKRKPSWTDRVLYKVVANNYENVTLRADLVSYNHMPHYTLSDHKPVVAQFNIKAFANYTEKTVEFAPINRVWYIGDADFRTQCTLTPDIEVNPGDWIGIYDANFHNLDDYISYEYLAKVGLSVRPASDGQPRTITLSFPVGSGVRTPGFYRFLYFSQPNSDVRSVLGISEPFEVSSKEDKLVTLGTSEEPSTSSSPGQPKSATASTDIFNDFTGLDVAKLSRHFSNDLSID
Summary
Uniprot
A0A3S2LQE0
A0A0N1ICS9
A0A194PY49
A0A2J7QUS7
A0A2J7QUS5
A0A2J7QUS6
+ More
A0A0L7L0J8 A0A067QIQ3 D7EHS9 A0A1Y1M983 A0A0C9PVF6 A0A023F059 A0A1Y1M7I0 A0A232FFI3 K7IMQ7 A0A2J7QUS8 U4UD51 A0A0C9QXM2 A0A146M744 A0A0A9YBR6 A0A2A3E7H1 A0A087ZR40 A0A154PHK8 A0A2P8ZJ26 A0A1W4WZT6 E0V9K5 A0A0P4VV98 A0A0N0U7K0 A0A0L7QJY7 A0A1W4X0U3 A0A084VK89 Q7PYY6 A0A1Q3G1H0 A0A2M4BN42 A0A1Q3G1G6 A0A1L8E5G5 A0A1Q3G1L9 A0A1L8E567 A0A2M3Z863 A0A2M4BN36 A0A1Q3G1H3 A0A1Q3G1H5 A0A1Q3G1E6 W5JTG5 A0A1B0CN17 A0A2M4AJE3 A0A2M4AKJ7 A0A2M4CWM3 A0A2M4CVU3 A0A2M4CWJ3 A0A026X2W7 Q17N17 A0A2M3Z7K2 A0A2M3Z7T2 A0A2M4CXW6 E2BJT2 A0A310SXJ3 A0A158NG32 A0A151I1F1 A0A151XBD6 F4WTM5 A0A195F239 A0A226EML7 A0A195E723 A0A1L8E592 A0A1L8E4Y5 A0A195CHY4 A0A182VCV1 A0A336KR75 A0A336MAV0 E9J5P1 A0A1D2N8T5 B0WM64 A0A2R7WX66 A0A182MXK2 A0A1J1HXG7 A0A2P6KMW4 A0A087T6S0 E2AS19 A0A182H053 A0A1B0DNX7 A0A131XA54 A0A0K8TLZ0 A0A131YMI8 A0A2H8TQ97 A0A131YPT9 A0A293MAA0
A0A0L7L0J8 A0A067QIQ3 D7EHS9 A0A1Y1M983 A0A0C9PVF6 A0A023F059 A0A1Y1M7I0 A0A232FFI3 K7IMQ7 A0A2J7QUS8 U4UD51 A0A0C9QXM2 A0A146M744 A0A0A9YBR6 A0A2A3E7H1 A0A087ZR40 A0A154PHK8 A0A2P8ZJ26 A0A1W4WZT6 E0V9K5 A0A0P4VV98 A0A0N0U7K0 A0A0L7QJY7 A0A1W4X0U3 A0A084VK89 Q7PYY6 A0A1Q3G1H0 A0A2M4BN42 A0A1Q3G1G6 A0A1L8E5G5 A0A1Q3G1L9 A0A1L8E567 A0A2M3Z863 A0A2M4BN36 A0A1Q3G1H3 A0A1Q3G1H5 A0A1Q3G1E6 W5JTG5 A0A1B0CN17 A0A2M4AJE3 A0A2M4AKJ7 A0A2M4CWM3 A0A2M4CVU3 A0A2M4CWJ3 A0A026X2W7 Q17N17 A0A2M3Z7K2 A0A2M3Z7T2 A0A2M4CXW6 E2BJT2 A0A310SXJ3 A0A158NG32 A0A151I1F1 A0A151XBD6 F4WTM5 A0A195F239 A0A226EML7 A0A195E723 A0A1L8E592 A0A1L8E4Y5 A0A195CHY4 A0A182VCV1 A0A336KR75 A0A336MAV0 E9J5P1 A0A1D2N8T5 B0WM64 A0A2R7WX66 A0A182MXK2 A0A1J1HXG7 A0A2P6KMW4 A0A087T6S0 E2AS19 A0A182H053 A0A1B0DNX7 A0A131XA54 A0A0K8TLZ0 A0A131YMI8 A0A2H8TQ97 A0A131YPT9 A0A293MAA0
Pubmed
26354079
26227816
24845553
18362917
19820115
28004739
+ More
25474469 28648823 20075255 23537049 26823975 25401762 29403074 20566863 27129103 24438588 12364791 14747013 17210077 20920257 23761445 24508170 30249741 17510324 20798317 21347285 21719571 21282665 27289101 26483478 28049606 26369729 26830274
25474469 28648823 20075255 23537049 26823975 25401762 29403074 20566863 27129103 24438588 12364791 14747013 17210077 20920257 23761445 24508170 30249741 17510324 20798317 21347285 21719571 21282665 27289101 26483478 28049606 26369729 26830274
EMBL
RSAL01000030
RVE51680.1
KQ459761
KPJ20134.1
KQ459589
KPI97674.1
+ More
NEVH01010486 PNF32341.1 PNF32339.1 PNF32338.1 JTDY01003776 KOB69023.1 KK853497 KDR07202.1 DS497665 EFA12122.1 GEZM01042397 JAV79887.1 GBYB01005423 JAG75190.1 GBBI01003832 JAC14880.1 GEZM01042398 JAV79886.1 NNAY01000314 OXU29280.1 PNF32340.1 KB632070 ERL88526.1 GBYB01005422 JAG75189.1 GDHC01004427 GDHC01003051 JAQ14202.1 JAQ15578.1 GBHO01013077 JAG30527.1 KZ288344 PBC27713.1 KQ434893 KZC10680.1 PYGN01000041 PSN56506.1 DS234994 EEB10061.1 GDKW01003050 JAI53545.1 KQ435709 KOX79978.1 KQ415017 KOC58938.1 ATLV01014160 KE524946 KFB38383.1 AAAB01008986 EAA00417.4 GFDL01001391 JAV33654.1 GGFJ01005359 MBW54500.1 GFDL01001400 JAV33645.1 GFDF01000299 JAV13785.1 GFDL01001364 JAV33681.1 GFDF01000300 JAV13784.1 GGFM01003972 MBW24723.1 GGFJ01005358 MBW54499.1 GFDL01001393 JAV33652.1 GFDL01001414 JAV33631.1 GFDL01001423 JAV33622.1 ADMH02000194 ETN67411.1 AJWK01019622 AJWK01019623 GGFK01007589 MBW40910.1 GGFK01007995 MBW41316.1 GGFL01005433 MBW69611.1 GGFL01005275 MBW69453.1 GGFL01005526 MBW69704.1 KK107020 QOIP01000004 EZA62453.1 RLU23804.1 CH477202 EAT48128.1 GGFM01003755 MBW24506.1 GGFM01003835 MBW24586.1 GGFL01005530 MBW69708.1 GL448636 EFN84057.1 KQ759804 OAD62806.1 ADTU01014952 KQ976580 KYM79940.1 KQ982335 KYQ57644.1 GL888341 EGI62446.1 KQ981864 KYN34159.1 LNIX01000003 OXA58377.1 KQ979568 KYN20988.1 GFDF01000295 JAV13789.1 GFDF01000294 JAV13790.1 KQ977721 KYN00350.1 UFQS01000631 UFQT01000631 SSX05553.1 SSX25912.1 SSX05554.1 SSX25913.1 GL768174 EFZ11865.1 LJIJ01000145 ODN01654.1 DS231995 EDS30898.1 KK855855 PTY24134.1 CVRI01000020 CRK90833.1 MWRG01008391 PRD27690.1 KK113694 KFM60809.1 GL442209 EFN63793.1 JXUM01023107 JXUM01023108 KQ560651 KXJ81442.1 AJVK01007816 AJVK01007817 AJVK01007818 AJVK01007819 AJVK01007820 GEFH01004668 JAP63913.1 GDAI01002653 JAI14950.1 GEDV01008098 JAP80459.1 GFXV01004569 MBW16374.1 GEDV01008097 JAP80460.1 GFWV01012995 MAA37724.1
NEVH01010486 PNF32341.1 PNF32339.1 PNF32338.1 JTDY01003776 KOB69023.1 KK853497 KDR07202.1 DS497665 EFA12122.1 GEZM01042397 JAV79887.1 GBYB01005423 JAG75190.1 GBBI01003832 JAC14880.1 GEZM01042398 JAV79886.1 NNAY01000314 OXU29280.1 PNF32340.1 KB632070 ERL88526.1 GBYB01005422 JAG75189.1 GDHC01004427 GDHC01003051 JAQ14202.1 JAQ15578.1 GBHO01013077 JAG30527.1 KZ288344 PBC27713.1 KQ434893 KZC10680.1 PYGN01000041 PSN56506.1 DS234994 EEB10061.1 GDKW01003050 JAI53545.1 KQ435709 KOX79978.1 KQ415017 KOC58938.1 ATLV01014160 KE524946 KFB38383.1 AAAB01008986 EAA00417.4 GFDL01001391 JAV33654.1 GGFJ01005359 MBW54500.1 GFDL01001400 JAV33645.1 GFDF01000299 JAV13785.1 GFDL01001364 JAV33681.1 GFDF01000300 JAV13784.1 GGFM01003972 MBW24723.1 GGFJ01005358 MBW54499.1 GFDL01001393 JAV33652.1 GFDL01001414 JAV33631.1 GFDL01001423 JAV33622.1 ADMH02000194 ETN67411.1 AJWK01019622 AJWK01019623 GGFK01007589 MBW40910.1 GGFK01007995 MBW41316.1 GGFL01005433 MBW69611.1 GGFL01005275 MBW69453.1 GGFL01005526 MBW69704.1 KK107020 QOIP01000004 EZA62453.1 RLU23804.1 CH477202 EAT48128.1 GGFM01003755 MBW24506.1 GGFM01003835 MBW24586.1 GGFL01005530 MBW69708.1 GL448636 EFN84057.1 KQ759804 OAD62806.1 ADTU01014952 KQ976580 KYM79940.1 KQ982335 KYQ57644.1 GL888341 EGI62446.1 KQ981864 KYN34159.1 LNIX01000003 OXA58377.1 KQ979568 KYN20988.1 GFDF01000295 JAV13789.1 GFDF01000294 JAV13790.1 KQ977721 KYN00350.1 UFQS01000631 UFQT01000631 SSX05553.1 SSX25912.1 SSX05554.1 SSX25913.1 GL768174 EFZ11865.1 LJIJ01000145 ODN01654.1 DS231995 EDS30898.1 KK855855 PTY24134.1 CVRI01000020 CRK90833.1 MWRG01008391 PRD27690.1 KK113694 KFM60809.1 GL442209 EFN63793.1 JXUM01023107 JXUM01023108 KQ560651 KXJ81442.1 AJVK01007816 AJVK01007817 AJVK01007818 AJVK01007819 AJVK01007820 GEFH01004668 JAP63913.1 GDAI01002653 JAI14950.1 GEDV01008098 JAP80459.1 GFXV01004569 MBW16374.1 GEDV01008097 JAP80460.1 GFWV01012995 MAA37724.1
Proteomes
UP000283053
UP000053240
UP000053268
UP000235965
UP000037510
UP000027135
+ More
UP000007266 UP000215335 UP000002358 UP000030742 UP000242457 UP000005203 UP000076502 UP000245037 UP000192223 UP000009046 UP000053105 UP000053825 UP000030765 UP000007062 UP000000673 UP000092461 UP000053097 UP000279307 UP000008820 UP000008237 UP000005205 UP000078540 UP000075809 UP000007755 UP000078541 UP000198287 UP000078492 UP000078542 UP000075903 UP000094527 UP000002320 UP000075884 UP000183832 UP000054359 UP000000311 UP000069940 UP000249989 UP000092462
UP000007266 UP000215335 UP000002358 UP000030742 UP000242457 UP000005203 UP000076502 UP000245037 UP000192223 UP000009046 UP000053105 UP000053825 UP000030765 UP000007062 UP000000673 UP000092461 UP000053097 UP000279307 UP000008820 UP000008237 UP000005205 UP000078540 UP000075809 UP000007755 UP000078541 UP000198287 UP000078492 UP000078542 UP000075903 UP000094527 UP000002320 UP000075884 UP000183832 UP000054359 UP000000311 UP000069940 UP000249989 UP000092462
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
A0A3S2LQE0
A0A0N1ICS9
A0A194PY49
A0A2J7QUS7
A0A2J7QUS5
A0A2J7QUS6
+ More
A0A0L7L0J8 A0A067QIQ3 D7EHS9 A0A1Y1M983 A0A0C9PVF6 A0A023F059 A0A1Y1M7I0 A0A232FFI3 K7IMQ7 A0A2J7QUS8 U4UD51 A0A0C9QXM2 A0A146M744 A0A0A9YBR6 A0A2A3E7H1 A0A087ZR40 A0A154PHK8 A0A2P8ZJ26 A0A1W4WZT6 E0V9K5 A0A0P4VV98 A0A0N0U7K0 A0A0L7QJY7 A0A1W4X0U3 A0A084VK89 Q7PYY6 A0A1Q3G1H0 A0A2M4BN42 A0A1Q3G1G6 A0A1L8E5G5 A0A1Q3G1L9 A0A1L8E567 A0A2M3Z863 A0A2M4BN36 A0A1Q3G1H3 A0A1Q3G1H5 A0A1Q3G1E6 W5JTG5 A0A1B0CN17 A0A2M4AJE3 A0A2M4AKJ7 A0A2M4CWM3 A0A2M4CVU3 A0A2M4CWJ3 A0A026X2W7 Q17N17 A0A2M3Z7K2 A0A2M3Z7T2 A0A2M4CXW6 E2BJT2 A0A310SXJ3 A0A158NG32 A0A151I1F1 A0A151XBD6 F4WTM5 A0A195F239 A0A226EML7 A0A195E723 A0A1L8E592 A0A1L8E4Y5 A0A195CHY4 A0A182VCV1 A0A336KR75 A0A336MAV0 E9J5P1 A0A1D2N8T5 B0WM64 A0A2R7WX66 A0A182MXK2 A0A1J1HXG7 A0A2P6KMW4 A0A087T6S0 E2AS19 A0A182H053 A0A1B0DNX7 A0A131XA54 A0A0K8TLZ0 A0A131YMI8 A0A2H8TQ97 A0A131YPT9 A0A293MAA0
A0A0L7L0J8 A0A067QIQ3 D7EHS9 A0A1Y1M983 A0A0C9PVF6 A0A023F059 A0A1Y1M7I0 A0A232FFI3 K7IMQ7 A0A2J7QUS8 U4UD51 A0A0C9QXM2 A0A146M744 A0A0A9YBR6 A0A2A3E7H1 A0A087ZR40 A0A154PHK8 A0A2P8ZJ26 A0A1W4WZT6 E0V9K5 A0A0P4VV98 A0A0N0U7K0 A0A0L7QJY7 A0A1W4X0U3 A0A084VK89 Q7PYY6 A0A1Q3G1H0 A0A2M4BN42 A0A1Q3G1G6 A0A1L8E5G5 A0A1Q3G1L9 A0A1L8E567 A0A2M3Z863 A0A2M4BN36 A0A1Q3G1H3 A0A1Q3G1H5 A0A1Q3G1E6 W5JTG5 A0A1B0CN17 A0A2M4AJE3 A0A2M4AKJ7 A0A2M4CWM3 A0A2M4CVU3 A0A2M4CWJ3 A0A026X2W7 Q17N17 A0A2M3Z7K2 A0A2M3Z7T2 A0A2M4CXW6 E2BJT2 A0A310SXJ3 A0A158NG32 A0A151I1F1 A0A151XBD6 F4WTM5 A0A195F239 A0A226EML7 A0A195E723 A0A1L8E592 A0A1L8E4Y5 A0A195CHY4 A0A182VCV1 A0A336KR75 A0A336MAV0 E9J5P1 A0A1D2N8T5 B0WM64 A0A2R7WX66 A0A182MXK2 A0A1J1HXG7 A0A2P6KMW4 A0A087T6S0 E2AS19 A0A182H053 A0A1B0DNX7 A0A131XA54 A0A0K8TLZ0 A0A131YMI8 A0A2H8TQ97 A0A131YPT9 A0A293MAA0
PDB
1I9Z
E-value=8.04948e-40,
Score=412
Ontologies
PATHWAY
GO
Topology
Subcellular location
Nucleus
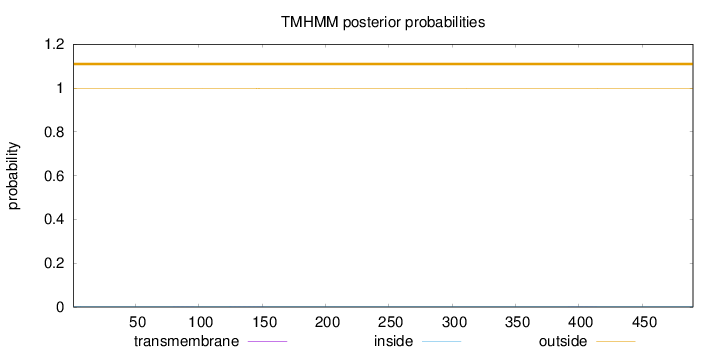
Length:
490
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03058
Exp number, first 60 AAs:
0.00042
Total prob of N-in:
0.00310
outside
1 - 490
Population Genetic Test Statistics
Pi
265.611808
Theta
18.645013
Tajima's D
1.261829
CLR
1.033734
CSRT
0.734313284335783
Interpretation
Uncertain
