Gene
KWMTBOMO04405
Annotation
PREDICTED:_G-protein_coupled_receptor_Mth2-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.934
Sequence
CDS
ATGGAAACTTTTTGGAACGAAACCTATCCGGCTGGGGTAACATTGCCCATGGTTTGCTTCACTACGCCTTTAGAAGAAACTAAGACTTCTGCCACTCTCATCGTTTATGCTACAGGACTTTTGATATCTGTACCGTTTTTACTGGCAACAGCATGTGTGTATCTATGTATATGCGAGCTTAGAGACACACACGGCAAGGCTCTGGCGTGCCATACAATCTGTCTGGCGTTGGCATTCATATGTTTGGCGACAACACAGTTGGCGGGCCATGCTTTTCCTAGCACTGCCTGTAATGTTATGGCGTACATCATCCAGTTCTCATTTGTCTCATGTTTTTTCTGGTTGAATGTGATGTGCTTCGATATATTTTTAAATGTTAGGAGATACGTAAATTCATCAGTAACCAGAAGAAGCATGAGACGCCGTTTTGCCTGGTACTGCGCGTATGCAGTACTGCTACCAGTATTACTGTTGATCATCACGGTCACAATGGACTTATCGCCGGCTGTGCCAAGCACGTACCTCAAGCCCAACTTCGGAGTAGTGGGGTGCTGGTTTAAAACGGATCAAGCAGCACTGCCATATTTCTATGGGCCGGTAGCCGTGATTTTGTTCAGCAACTTAATTATATTTTTCCTAACTTCTCGAGCCTTTGCCAAGCATTACGACAAGCTGAAAGACCTGACTCCGTTAGATATGGCACATTCAGACTTCAGTACATCAGATGACTGGGTCAGGTTGAGTGCAGTACTGGGGCAGCCCGTTACGAATCAGGAGCAAACATCACCCAGAGAAGCGCCCACTAACTTTAGTCAGAGATTGAAAAAGTACAAGAAGATTTTCCGAACATGCTGTATGCTTTCCTTCATCATGGGTCTATCATGGGTGCTAGAGGTGGTCTCCTGGGCGGCTGGATCTGGTTCCACCGCGGTGTCGGTGTGGTCCATCTTCGATCTCATCAACGCTCTCCAAGGAGTAATGATCTTTGCCATTTTCGTGTTGCAGCAACCCGTTCGATCTTATGTTAAGTCTTCAAAATTCTTCTCAACGTGTCGAAGTAAAAGGAACAGCTCCGAGGTTTCAGTTGTGAGTGAATGGAATAGCAATCCAGGACAAGGAAGAAGAGACTATGTATAG
Protein
METFWNETYPAGVTLPMVCFTTPLEETKTSATLIVYATGLLISVPFLLATACVYLCICELRDTHGKALACHTICLALAFICLATTQLAGHAFPSTACNVMAYIIQFSFVSCFFWLNVMCFDIFLNVRRYVNSSVTRRSMRRRFAWYCAYAVLLPVLLLIITVTMDLSPAVPSTYLKPNFGVVGCWFKTDQAALPYFYGPVAVILFSNLIIFFLTSRAFAKHYDKLKDLTPLDMAHSDFSTSDDWVRLSAVLGQPVTNQEQTSPREAPTNFSQRLKKYKKIFRTCCMLSFIMGLSWVLEVVSWAAGSGSTAVSVWSIFDLINALQGVMIFAIFVLQQPVRSYVKSSKFFSTCRSKRNSSEVSVVSEWNSNPGQGRRDYV
Summary
Uniprot
A0A2H1WU47
A0A1E1WU24
A0A212FF91
A0A0N0PEM5
A0A194PWH6
A0A3S2PHL1
+ More
E2BJT1 K7IMQ6 A0A067QGP0 A0A0C9R4E3 E9J5P0 A0A026X2D5 A0A232FFQ2 A0A3L8DUG5 A0A151I182 A0A158NG34 A0A0C9R2H1 A0A195CI13 F4WTM4 A0A195F0Q2 A0A195E835 A0A151XB81 A0A1B6CNV6 A0A1B6KGW1 A0A1B6EP37 E2AS18 A0A154PFR1 A0A0L7QK49 A0A2J7QUS9 A0A2J7R7H5 A0A023F6G4 A0A1B6K827 A0A310SYI0 A0A2A3E881 A0A1B6DQS8 K7JQP3 J9JKN2 A0A1B6F3W1 A0A1B6JEQ5 A0A1B6KQY4 A0A1B6G5F1 A0A1B6LT28 A0A1B6F351 A0A154NYM6 A0A067QSF7 A0A2H8TDW7 A0A1B6IBM7 A0A1B6IUT9 A0A1B6JBW8 A0A1E1WH08 A0A1E1WCB0 A0A026WB41 A0A1E1WN64 J7ETA9 E0VM50 A0A1B6CU80 F4WY56 A0A1B6E0C8 A0A151X6P0 A0A0K8S4J2 A0A3L8D8G7 A0A0M9ABW3 A0A195CCU5 A0A0N0BJ15 A0A1B6KT72 R4FKF4 A0A087ZR19 A0A151I3Y4 A0A195EHH8 A0A158NSZ5 A0A0L7QS61 A0A195FEV0 E2AK97 A0A195EU48 A0A2S2QB80 A0A1B6EGM6 E2AKW8 A0A2R7WXZ9 A0A158NGK2 A0A195BA00 A0A212FD97 A0A195E5N6 A0A0P5Q5G3 A0A146MAI8 A0A0A9YUT1 A0A182N958 A0A0A9Z0N2 A0A1Y1KDY1 A0A2Y9D3M1 A0A1Z1LVU5 Q7PNT9 A0A0P5IYH2 A0A0P5K667 A0A0P5E0J6 A0A0P5ECE8 A0A182LLL5 A0A182TDZ9
E2BJT1 K7IMQ6 A0A067QGP0 A0A0C9R4E3 E9J5P0 A0A026X2D5 A0A232FFQ2 A0A3L8DUG5 A0A151I182 A0A158NG34 A0A0C9R2H1 A0A195CI13 F4WTM4 A0A195F0Q2 A0A195E835 A0A151XB81 A0A1B6CNV6 A0A1B6KGW1 A0A1B6EP37 E2AS18 A0A154PFR1 A0A0L7QK49 A0A2J7QUS9 A0A2J7R7H5 A0A023F6G4 A0A1B6K827 A0A310SYI0 A0A2A3E881 A0A1B6DQS8 K7JQP3 J9JKN2 A0A1B6F3W1 A0A1B6JEQ5 A0A1B6KQY4 A0A1B6G5F1 A0A1B6LT28 A0A1B6F351 A0A154NYM6 A0A067QSF7 A0A2H8TDW7 A0A1B6IBM7 A0A1B6IUT9 A0A1B6JBW8 A0A1E1WH08 A0A1E1WCB0 A0A026WB41 A0A1E1WN64 J7ETA9 E0VM50 A0A1B6CU80 F4WY56 A0A1B6E0C8 A0A151X6P0 A0A0K8S4J2 A0A3L8D8G7 A0A0M9ABW3 A0A195CCU5 A0A0N0BJ15 A0A1B6KT72 R4FKF4 A0A087ZR19 A0A151I3Y4 A0A195EHH8 A0A158NSZ5 A0A0L7QS61 A0A195FEV0 E2AK97 A0A195EU48 A0A2S2QB80 A0A1B6EGM6 E2AKW8 A0A2R7WXZ9 A0A158NGK2 A0A195BA00 A0A212FD97 A0A195E5N6 A0A0P5Q5G3 A0A146MAI8 A0A0A9YUT1 A0A182N958 A0A0A9Z0N2 A0A1Y1KDY1 A0A2Y9D3M1 A0A1Z1LVU5 Q7PNT9 A0A0P5IYH2 A0A0P5K667 A0A0P5E0J6 A0A0P5ECE8 A0A182LLL5 A0A182TDZ9
Pubmed
EMBL
ODYU01011040
SOQ56527.1
GDQN01000708
JAT90346.1
AGBW02008846
OWR52401.1
+ More
KQ459761 KPJ20136.1 KQ459589 KPI97672.1 RSAL01000030 RVE51681.1 GL448636 EFN84056.1 KK853497 KDR07201.1 GBYB01001691 JAG71458.1 GL768174 EFZ11847.1 KK107020 EZA62452.1 NNAY01000314 OXU29288.1 QOIP01000004 RLU23813.1 KQ976580 KYM79939.1 ADTU01014952 GBYB01010274 JAG80041.1 KQ977721 KYN00348.1 GL888341 EGI62445.1 KQ981864 KYN34160.1 KQ979568 KYN20987.1 KQ982335 KYQ57643.1 GEDC01022190 JAS15108.1 GEBQ01029296 GEBQ01011028 JAT10681.1 JAT28949.1 GECZ01030063 JAS39706.1 GL442209 EFN63792.1 KQ434893 KZC10681.1 KQ415017 KOC58935.1 NEVH01010486 PNF32336.1 NEVH01006732 PNF36788.1 GBBI01001888 JAC16824.1 GECU01000107 JAT07600.1 KQ759804 OAD62804.1 KZ288344 PBC27714.1 GEDC01018773 GEDC01009262 GEDC01000043 JAS18525.1 JAS28036.1 JAS37255.1 ABLF02038161 GECZ01024998 JAS44771.1 GECU01010087 JAS97619.1 GEBQ01026124 JAT13853.1 GECZ01012105 JAS57664.1 GEBQ01013134 JAT26843.1 GECZ01025080 JAS44689.1 KQ434783 KZC04727.1 KK853008 KDR12560.1 GFXV01000486 MBW12291.1 GECU01023436 JAS84270.1 GECU01017005 JAS90701.1 GECU01011109 GECU01002381 JAS96597.1 JAT05326.1 GDQN01004782 JAT86272.1 GDQN01006455 JAT84599.1 KK107309 EZA52891.1 GDQN01002620 JAT88434.1 HQ188200 AEM43035.1 DS235289 EEB14456.1 GEDC01020320 JAS16978.1 GL888439 EGI60813.1 GEDC01018212 GEDC01005937 GEDC01001428 JAS19086.1 JAS31361.1 JAS35870.1 KQ982476 KYQ56046.1 GBRD01017616 JAG48211.1 QOIP01000011 RLU16797.1 KQ435709 KOX79979.1 KQ978023 KYM98041.1 KQ435724 KOX78511.1 GEBQ01025533 JAT14444.1 GAHY01001873 JAA75637.1 KQ976472 KYM83995.1 KQ978957 KYN27327.1 ADTU01025461 ADTU01025462 KQ414758 KOC61468.1 KQ981636 KYN38891.1 GL440209 EFN66138.1 KQ981965 KYN31773.1 GGMS01005791 MBY74994.1 GEDC01000202 JAS37096.1 GL440425 EFN65885.1 KK855935 PTY24417.1 ADTU01015065 KQ976542 KYM81024.1 AGBW02009085 OWR51711.1 KQ979608 KYN20401.1 GDIQ01119126 JAL32600.1 GDHC01001866 JAQ16763.1 GBHO01008233 JAG35371.1 GBHO01008234 JAG35370.1 GEZM01086187 JAV59692.1 KX009474 ARW56807.1 AAAB01008960 EAA11607.3 GDIQ01251881 GDIQ01250437 GDIQ01207654 GDIQ01206244 GDIQ01188398 JAK44071.1 GDIQ01251880 GDIQ01250436 GDIQ01208952 GDIQ01207653 GDIQ01206246 GDIQ01190154 JAK61571.1 GDIP01149638 JAJ73764.1 GDIP01143981 JAJ79421.1
KQ459761 KPJ20136.1 KQ459589 KPI97672.1 RSAL01000030 RVE51681.1 GL448636 EFN84056.1 KK853497 KDR07201.1 GBYB01001691 JAG71458.1 GL768174 EFZ11847.1 KK107020 EZA62452.1 NNAY01000314 OXU29288.1 QOIP01000004 RLU23813.1 KQ976580 KYM79939.1 ADTU01014952 GBYB01010274 JAG80041.1 KQ977721 KYN00348.1 GL888341 EGI62445.1 KQ981864 KYN34160.1 KQ979568 KYN20987.1 KQ982335 KYQ57643.1 GEDC01022190 JAS15108.1 GEBQ01029296 GEBQ01011028 JAT10681.1 JAT28949.1 GECZ01030063 JAS39706.1 GL442209 EFN63792.1 KQ434893 KZC10681.1 KQ415017 KOC58935.1 NEVH01010486 PNF32336.1 NEVH01006732 PNF36788.1 GBBI01001888 JAC16824.1 GECU01000107 JAT07600.1 KQ759804 OAD62804.1 KZ288344 PBC27714.1 GEDC01018773 GEDC01009262 GEDC01000043 JAS18525.1 JAS28036.1 JAS37255.1 ABLF02038161 GECZ01024998 JAS44771.1 GECU01010087 JAS97619.1 GEBQ01026124 JAT13853.1 GECZ01012105 JAS57664.1 GEBQ01013134 JAT26843.1 GECZ01025080 JAS44689.1 KQ434783 KZC04727.1 KK853008 KDR12560.1 GFXV01000486 MBW12291.1 GECU01023436 JAS84270.1 GECU01017005 JAS90701.1 GECU01011109 GECU01002381 JAS96597.1 JAT05326.1 GDQN01004782 JAT86272.1 GDQN01006455 JAT84599.1 KK107309 EZA52891.1 GDQN01002620 JAT88434.1 HQ188200 AEM43035.1 DS235289 EEB14456.1 GEDC01020320 JAS16978.1 GL888439 EGI60813.1 GEDC01018212 GEDC01005937 GEDC01001428 JAS19086.1 JAS31361.1 JAS35870.1 KQ982476 KYQ56046.1 GBRD01017616 JAG48211.1 QOIP01000011 RLU16797.1 KQ435709 KOX79979.1 KQ978023 KYM98041.1 KQ435724 KOX78511.1 GEBQ01025533 JAT14444.1 GAHY01001873 JAA75637.1 KQ976472 KYM83995.1 KQ978957 KYN27327.1 ADTU01025461 ADTU01025462 KQ414758 KOC61468.1 KQ981636 KYN38891.1 GL440209 EFN66138.1 KQ981965 KYN31773.1 GGMS01005791 MBY74994.1 GEDC01000202 JAS37096.1 GL440425 EFN65885.1 KK855935 PTY24417.1 ADTU01015065 KQ976542 KYM81024.1 AGBW02009085 OWR51711.1 KQ979608 KYN20401.1 GDIQ01119126 JAL32600.1 GDHC01001866 JAQ16763.1 GBHO01008233 JAG35371.1 GBHO01008234 JAG35370.1 GEZM01086187 JAV59692.1 KX009474 ARW56807.1 AAAB01008960 EAA11607.3 GDIQ01251881 GDIQ01250437 GDIQ01207654 GDIQ01206244 GDIQ01188398 JAK44071.1 GDIQ01251880 GDIQ01250436 GDIQ01208952 GDIQ01207653 GDIQ01206246 GDIQ01190154 JAK61571.1 GDIP01149638 JAJ73764.1 GDIP01143981 JAJ79421.1
Proteomes
UP000007151
UP000053240
UP000053268
UP000283053
UP000008237
UP000002358
+ More
UP000027135 UP000053097 UP000215335 UP000279307 UP000078540 UP000005205 UP000078542 UP000007755 UP000078541 UP000078492 UP000075809 UP000000311 UP000076502 UP000053825 UP000235965 UP000242457 UP000007819 UP000005203 UP000009046 UP000053105 UP000075884 UP000076407 UP000007062 UP000075882 UP000075902
UP000027135 UP000053097 UP000215335 UP000279307 UP000078540 UP000005205 UP000078542 UP000007755 UP000078541 UP000078492 UP000075809 UP000000311 UP000076502 UP000053825 UP000235965 UP000242457 UP000007819 UP000005203 UP000009046 UP000053105 UP000075884 UP000076407 UP000007062 UP000075882 UP000075902
Interpro
SUPFAM
SSF63877
SSF63877
Gene 3D
CDD
ProteinModelPortal
A0A2H1WU47
A0A1E1WU24
A0A212FF91
A0A0N0PEM5
A0A194PWH6
A0A3S2PHL1
+ More
E2BJT1 K7IMQ6 A0A067QGP0 A0A0C9R4E3 E9J5P0 A0A026X2D5 A0A232FFQ2 A0A3L8DUG5 A0A151I182 A0A158NG34 A0A0C9R2H1 A0A195CI13 F4WTM4 A0A195F0Q2 A0A195E835 A0A151XB81 A0A1B6CNV6 A0A1B6KGW1 A0A1B6EP37 E2AS18 A0A154PFR1 A0A0L7QK49 A0A2J7QUS9 A0A2J7R7H5 A0A023F6G4 A0A1B6K827 A0A310SYI0 A0A2A3E881 A0A1B6DQS8 K7JQP3 J9JKN2 A0A1B6F3W1 A0A1B6JEQ5 A0A1B6KQY4 A0A1B6G5F1 A0A1B6LT28 A0A1B6F351 A0A154NYM6 A0A067QSF7 A0A2H8TDW7 A0A1B6IBM7 A0A1B6IUT9 A0A1B6JBW8 A0A1E1WH08 A0A1E1WCB0 A0A026WB41 A0A1E1WN64 J7ETA9 E0VM50 A0A1B6CU80 F4WY56 A0A1B6E0C8 A0A151X6P0 A0A0K8S4J2 A0A3L8D8G7 A0A0M9ABW3 A0A195CCU5 A0A0N0BJ15 A0A1B6KT72 R4FKF4 A0A087ZR19 A0A151I3Y4 A0A195EHH8 A0A158NSZ5 A0A0L7QS61 A0A195FEV0 E2AK97 A0A195EU48 A0A2S2QB80 A0A1B6EGM6 E2AKW8 A0A2R7WXZ9 A0A158NGK2 A0A195BA00 A0A212FD97 A0A195E5N6 A0A0P5Q5G3 A0A146MAI8 A0A0A9YUT1 A0A182N958 A0A0A9Z0N2 A0A1Y1KDY1 A0A2Y9D3M1 A0A1Z1LVU5 Q7PNT9 A0A0P5IYH2 A0A0P5K667 A0A0P5E0J6 A0A0P5ECE8 A0A182LLL5 A0A182TDZ9
E2BJT1 K7IMQ6 A0A067QGP0 A0A0C9R4E3 E9J5P0 A0A026X2D5 A0A232FFQ2 A0A3L8DUG5 A0A151I182 A0A158NG34 A0A0C9R2H1 A0A195CI13 F4WTM4 A0A195F0Q2 A0A195E835 A0A151XB81 A0A1B6CNV6 A0A1B6KGW1 A0A1B6EP37 E2AS18 A0A154PFR1 A0A0L7QK49 A0A2J7QUS9 A0A2J7R7H5 A0A023F6G4 A0A1B6K827 A0A310SYI0 A0A2A3E881 A0A1B6DQS8 K7JQP3 J9JKN2 A0A1B6F3W1 A0A1B6JEQ5 A0A1B6KQY4 A0A1B6G5F1 A0A1B6LT28 A0A1B6F351 A0A154NYM6 A0A067QSF7 A0A2H8TDW7 A0A1B6IBM7 A0A1B6IUT9 A0A1B6JBW8 A0A1E1WH08 A0A1E1WCB0 A0A026WB41 A0A1E1WN64 J7ETA9 E0VM50 A0A1B6CU80 F4WY56 A0A1B6E0C8 A0A151X6P0 A0A0K8S4J2 A0A3L8D8G7 A0A0M9ABW3 A0A195CCU5 A0A0N0BJ15 A0A1B6KT72 R4FKF4 A0A087ZR19 A0A151I3Y4 A0A195EHH8 A0A158NSZ5 A0A0L7QS61 A0A195FEV0 E2AK97 A0A195EU48 A0A2S2QB80 A0A1B6EGM6 E2AKW8 A0A2R7WXZ9 A0A158NGK2 A0A195BA00 A0A212FD97 A0A195E5N6 A0A0P5Q5G3 A0A146MAI8 A0A0A9YUT1 A0A182N958 A0A0A9Z0N2 A0A1Y1KDY1 A0A2Y9D3M1 A0A1Z1LVU5 Q7PNT9 A0A0P5IYH2 A0A0P5K667 A0A0P5E0J6 A0A0P5ECE8 A0A182LLL5 A0A182TDZ9
Ontologies
GO
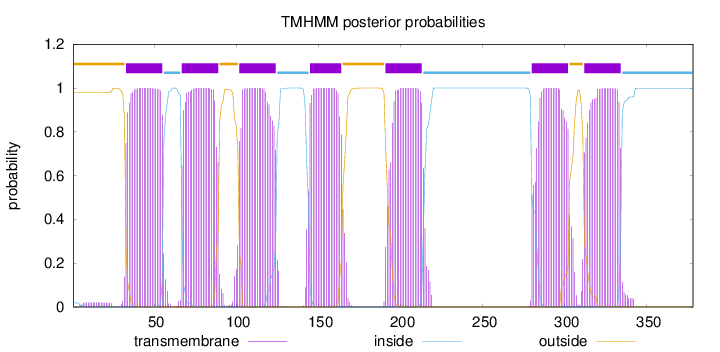
Topology
Length:
378
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
156.18868
Exp number, first 60 AAs:
23.39374
Total prob of N-in:
0.01985
POSSIBLE N-term signal
sequence
outside
1 - 32
TMhelix
33 - 55
inside
56 - 66
TMhelix
67 - 89
outside
90 - 101
TMhelix
102 - 124
inside
125 - 144
TMhelix
145 - 164
outside
165 - 190
TMhelix
191 - 213
inside
214 - 279
TMhelix
280 - 302
outside
303 - 311
TMhelix
312 - 334
inside
335 - 378
Population Genetic Test Statistics
Pi
239.063731
Theta
176.383068
Tajima's D
1.011977
CLR
0.050608
CSRT
0.664616769161542
Interpretation
Uncertain
