Gene
KWMTBOMO04404
Annotation
PREDICTED:_uncharacterized_protein_LOC101747137_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.711
Sequence
CDS
ATGGCGCAGGCGATAGCGAACGAATTGGACATCGCGGTCGGCAGAAGGACGTACTGGACTGACTCCAGTACAGTTCTGACATGGATAAAGACCGACCCACGCACGTTCAAACCTTTCGTTGCGCACCGACTCGCCGAGATAGAAGAGTCAACGAAGCCCCAAGAATGGCGATGGGTGCCTGGCTCGCAAAACCCAGCAGACGACGCAACTAGAGAGGCGCCGGCGGATTTCGACCACACGCATCGGTGGTTTAATGGACCCGAGTTCCTGACGTGGGATGAATCGCGCTGGCCCAAGCCGCGAACATTCAAGCAAGAACCGTCTGGAGAAGAGAAAGAGGCCTTTCTGGTCGCCACGGCGAGGACCGCTGACGCTAGACCCACGCCCGATCCGCACAGATTCTCGAGCTGGGTCAAGTTATTGAGAGCAACAGCCAGGGTCCTCCAATTTATCGAGCTGTGTCGACCCCGGAAGGAGAGCGCCTGCGTGTCCAGACAATTGGAGCAGCAAGATCCCACGTGGAGGACGACGCGAGCGAAGCAGCCCCGATCAACATGGAAGATAAGGACTCCAGAAGCACCTACAGAAGCGTGGCTGCCGCTCGATCCGCCCCACTTGAAGAAGGCGGAGAAGATCCTACTGAGAAGCAGCCAAGGAGAGAGCTTCGGTGAAAAAGATCCCGAACGTCACCCCAAGTTGCGACGGCTCGACGTCGTGATGGAAGATGGCCTGTTGCGCCTGCGCGGACGTATCGACGCAGCGCAATACATAGATGCAGGCTGCAAGCGACCGATCGTGCTGGACGGGAAGCACGTGATAGCGAGATTATTGATCAAACACTATCACGAGGCGTTCCAGCACGGTAACCACGCAACGGTAATTGTAAAAGTTCGCATTGTATAG
Protein
MAQAIANELDIAVGRRTYWTDSSTVLTWIKTDPRTFKPFVAHRLAEIEESTKPQEWRWVPGSQNPADDATREAPADFDHTHRWFNGPEFLTWDESRWPKPRTFKQEPSGEEKEAFLVATARTADARPTPDPHRFSSWVKLLRATARVLQFIELCRPRKESACVSRQLEQQDPTWRTTRAKQPRSTWKIRTPEAPTEAWLPLDPPHLKKAEKILLRSSQGESFGEKDPERHPKLRRLDVVMEDGLLRLRGRIDAAQYIDAGCKRPIVLDGKHVIARLLIKHYHEAFQHGNHATVIVKVRIV
Summary
Uniprot
Q2MGA5
Q9NDM9
Q9NDN0
A0A3S2N5J7
A0A2A4IRQ3
A0A0N1IGH3
+ More
A0A0L7LE26 A0A0L7KQM8 A0A226EFR1 K7JA41 A0A226DGY5 A0A0L7L8X1 A0A226D351 A0A226DH22 A0A226DM06 A0A226EKF8 A0A226EDU9 A0A182G7Z0 W8AJC8 A0A2H1VMV3 A0A182HCB4 A0A226E2P7 A0A1B6LK77 A0A2M4CKZ0 A0A182GL35 W8B882 W8AYL5 A0A2M4CKI9 A0A182G5I1 Q76C95 Q76C94 A0A2M4B9W9 A0A2M4B9D0 A0A182HZZ0 A0A182G158 O02007 A0A1W7R6H4 A0A182H4A2 A0A182HEF9 A0A226DIJ1 A0A0L7LJ73 A0A182H5B8 A0A3S2P6R4 A0A1W7R6M1 A0A2M4CVY8 A0A182GPV7 A0A226DTU1 A0A1W7R6F2 A0A146S4B8 A0A0S7G3I4 A0A2M4BC71 A0A2M4BC51 A0A146PP93 A0A1W7R681 A0A146QCZ7 A0A146S1B6 A0A146Q7X0 A0A0S7G7U6 A0A2M4B8T5 A0A2M4B8I4 A0A0S7FZ14 W4YU11 A0A146SP51 A0A034V9M7 A0A2B4RJC8 A0A1Y1LL65 A0A2G8KYS5 W4Y2R6 A0A0P5QIW7 A0A182GKP7 A0A182GRX9 A0A0P6C5E3 A0A1Y1LN76 A0A0P6AZF3 A0A2G8KVJ2 A0A1W7R6F7 A0A182YRW4 A0A2B4SHN3 A0A1B0CYJ8 A0A1S3QGM9 A0A146S2G4 A0A2B4SX75 A0A146S1A2 A0A146T9S8 A0A085N6I6 W4XIU5 G3NC29 W4XIU4 A0A1A8V6P1 W4XIU3 A0A0S7LK46 W4ZHM9 A0A1S3SW27 A0A146TZK4
A0A0L7LE26 A0A0L7KQM8 A0A226EFR1 K7JA41 A0A226DGY5 A0A0L7L8X1 A0A226D351 A0A226DH22 A0A226DM06 A0A226EKF8 A0A226EDU9 A0A182G7Z0 W8AJC8 A0A2H1VMV3 A0A182HCB4 A0A226E2P7 A0A1B6LK77 A0A2M4CKZ0 A0A182GL35 W8B882 W8AYL5 A0A2M4CKI9 A0A182G5I1 Q76C95 Q76C94 A0A2M4B9W9 A0A2M4B9D0 A0A182HZZ0 A0A182G158 O02007 A0A1W7R6H4 A0A182H4A2 A0A182HEF9 A0A226DIJ1 A0A0L7LJ73 A0A182H5B8 A0A3S2P6R4 A0A1W7R6M1 A0A2M4CVY8 A0A182GPV7 A0A226DTU1 A0A1W7R6F2 A0A146S4B8 A0A0S7G3I4 A0A2M4BC71 A0A2M4BC51 A0A146PP93 A0A1W7R681 A0A146QCZ7 A0A146S1B6 A0A146Q7X0 A0A0S7G7U6 A0A2M4B8T5 A0A2M4B8I4 A0A0S7FZ14 W4YU11 A0A146SP51 A0A034V9M7 A0A2B4RJC8 A0A1Y1LL65 A0A2G8KYS5 W4Y2R6 A0A0P5QIW7 A0A182GKP7 A0A182GRX9 A0A0P6C5E3 A0A1Y1LN76 A0A0P6AZF3 A0A2G8KVJ2 A0A1W7R6F7 A0A182YRW4 A0A2B4SHN3 A0A1B0CYJ8 A0A1S3QGM9 A0A146S2G4 A0A2B4SX75 A0A146S1A2 A0A146T9S8 A0A085N6I6 W4XIU5 G3NC29 W4XIU4 A0A1A8V6P1 W4XIU3 A0A0S7LK46 W4ZHM9 A0A1S3SW27 A0A146TZK4
Pubmed
EMBL
AF530470
AAQ09229.1
AB042119
BAA95570.1
AB042118
BAA95569.1
+ More
RSAL01000375 RVE42106.1 NWSH01008670 PCG62455.1 KQ459986 KPJ18861.1 JTDY01001530 KOB73634.1 JTDY01006922 KOB65592.1 LNIX01000004 OXA56382.1 AAZX01023278 LNIX01000019 OXA44812.1 JTDY01002261 KOB71779.1 LNIX01000039 OXA39294.1 OXA44855.1 LNIX01000015 OXA46575.1 LNIX01000003 OXA57597.1 OXA55408.1 JXUM01151198 KQ571164 KXJ68235.1 GAMC01017930 JAB88625.1 ODYU01003174 SOQ41584.1 JXUM01127084 KQ567147 KXJ69602.1 LNIX01000007 OXA52015.1 GEBQ01015876 JAT24101.1 GGFL01001842 MBW66020.1 JXUM01013493 KQ560378 KXJ82704.1 GAMC01020598 GAMC01020596 GAMC01020592 JAB85959.1 GAMC01020594 JAB85961.1 GGFL01001682 MBW65860.1 JXUM01146045 JXUM01146046 AB110069 BAD01589.1 BAD01590.1 GGFJ01000487 MBW49628.1 GGFJ01000471 MBW49612.1 APCN01002024 JXUM01136032 KQ568367 KXJ69026.1 D83207 BAA19772.1 GEHC01000914 JAV46731.1 JXUM01109099 KQ565290 KXJ71123.1 JXUM01005879 KQ560203 KXJ83865.1 LNIX01000018 OXA45362.1 JTDY01000902 KOB75497.1 JXUM01111221 KQ565482 KXJ70922.1 RSAL01000010 RVE53755.1 GEHC01000872 JAV46773.1 GGFL01005193 MBW69371.1 JXUM01016534 JXUM01056823 KQ561926 KQ560461 KXJ77135.1 KXJ82302.1 LNIX01000012 OXA48117.1 GEHC01000913 JAV46732.1 GCES01110894 JAQ75428.1 GBYX01459353 JAO22206.1 GGFJ01001476 MBW50617.1 GGFJ01001462 MBW50603.1 GCES01140631 JAQ45691.1 GEHC01000967 JAV46678.1 GCES01132273 JAQ54049.1 GCES01111900 JAQ74422.1 GCES01134385 JAQ51937.1 GBYX01459357 JAO22202.1 GGFJ01000302 MBW49443.1 GGFJ01000218 MBW49359.1 GBYX01459355 JAO22204.1 AAGJ04142769 AAGJ04142770 GCES01103975 JAQ82347.1 GAKP01020709 JAC38243.1 LSMT01000554 PFX16342.1 GEZM01052587 JAV74399.1 MRZV01000303 PIK53060.1 AAGJ04043389 GDIQ01114015 JAL37711.1 JXUM01013377 KQ560375 KXJ82728.1 JXUM01083542 KQ563383 KXJ73941.1 GDIP01004568 JAM99147.1 GEZM01052586 JAV74401.1 GDIP01036233 JAM67482.1 MRZV01000348 PIK51982.1 GEHC01000945 JAV46700.1 LSMT01000075 PFX28896.1 AJVK01001938 GCES01111762 JAQ74560.1 LSMT01000016 PFX33015.1 GCES01112266 JAQ74056.1 GCES01097166 JAQ89156.1 KL367545 KFD65082.1 AAGJ04001285 AAGJ04091218 HAEJ01014775 SBS55232.1 GBYX01164676 JAO89828.1 AAGJ04050745 GCES01088038 JAQ98284.1
RSAL01000375 RVE42106.1 NWSH01008670 PCG62455.1 KQ459986 KPJ18861.1 JTDY01001530 KOB73634.1 JTDY01006922 KOB65592.1 LNIX01000004 OXA56382.1 AAZX01023278 LNIX01000019 OXA44812.1 JTDY01002261 KOB71779.1 LNIX01000039 OXA39294.1 OXA44855.1 LNIX01000015 OXA46575.1 LNIX01000003 OXA57597.1 OXA55408.1 JXUM01151198 KQ571164 KXJ68235.1 GAMC01017930 JAB88625.1 ODYU01003174 SOQ41584.1 JXUM01127084 KQ567147 KXJ69602.1 LNIX01000007 OXA52015.1 GEBQ01015876 JAT24101.1 GGFL01001842 MBW66020.1 JXUM01013493 KQ560378 KXJ82704.1 GAMC01020598 GAMC01020596 GAMC01020592 JAB85959.1 GAMC01020594 JAB85961.1 GGFL01001682 MBW65860.1 JXUM01146045 JXUM01146046 AB110069 BAD01589.1 BAD01590.1 GGFJ01000487 MBW49628.1 GGFJ01000471 MBW49612.1 APCN01002024 JXUM01136032 KQ568367 KXJ69026.1 D83207 BAA19772.1 GEHC01000914 JAV46731.1 JXUM01109099 KQ565290 KXJ71123.1 JXUM01005879 KQ560203 KXJ83865.1 LNIX01000018 OXA45362.1 JTDY01000902 KOB75497.1 JXUM01111221 KQ565482 KXJ70922.1 RSAL01000010 RVE53755.1 GEHC01000872 JAV46773.1 GGFL01005193 MBW69371.1 JXUM01016534 JXUM01056823 KQ561926 KQ560461 KXJ77135.1 KXJ82302.1 LNIX01000012 OXA48117.1 GEHC01000913 JAV46732.1 GCES01110894 JAQ75428.1 GBYX01459353 JAO22206.1 GGFJ01001476 MBW50617.1 GGFJ01001462 MBW50603.1 GCES01140631 JAQ45691.1 GEHC01000967 JAV46678.1 GCES01132273 JAQ54049.1 GCES01111900 JAQ74422.1 GCES01134385 JAQ51937.1 GBYX01459357 JAO22202.1 GGFJ01000302 MBW49443.1 GGFJ01000218 MBW49359.1 GBYX01459355 JAO22204.1 AAGJ04142769 AAGJ04142770 GCES01103975 JAQ82347.1 GAKP01020709 JAC38243.1 LSMT01000554 PFX16342.1 GEZM01052587 JAV74399.1 MRZV01000303 PIK53060.1 AAGJ04043389 GDIQ01114015 JAL37711.1 JXUM01013377 KQ560375 KXJ82728.1 JXUM01083542 KQ563383 KXJ73941.1 GDIP01004568 JAM99147.1 GEZM01052586 JAV74401.1 GDIP01036233 JAM67482.1 MRZV01000348 PIK51982.1 GEHC01000945 JAV46700.1 LSMT01000075 PFX28896.1 AJVK01001938 GCES01111762 JAQ74560.1 LSMT01000016 PFX33015.1 GCES01112266 JAQ74056.1 GCES01097166 JAQ89156.1 KL367545 KFD65082.1 AAGJ04001285 AAGJ04091218 HAEJ01014775 SBS55232.1 GBYX01164676 JAO89828.1 AAGJ04050745 GCES01088038 JAQ98284.1
Proteomes
Pfam
Interpro
IPR040676
DUF5641
+ More
IPR036397 RNaseH_sf
IPR005312 DUF1759
IPR001584 Integrase_cat-core
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR041588 Integrase_H2C2
IPR008737 Peptidase_asp_put
IPR001995 Peptidase_A2_cat
IPR001611 Leu-rich_rpt
IPR032675 LRR_dom_sf
IPR001878 Znf_CCHC
IPR006094 Oxid_FAD_bind_N
IPR016166 FAD-bd_PCMH
IPR016164 FAD-linked_Oxase-like_C
IPR016170 Cytok_DH_C_sf
IPR015213 Cholesterol_OX_subst-bd
IPR036318 FAD-bd_PCMH-like_sf
IPR016169 FAD-bd_PCMH_sub2
IPR016167 FAD-bd_PCMH_sub1
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR000477 RT_dom
IPR019786 Zinc_finger_PHD-type_CS
IPR036875 Znf_CCHC_sf
IPR001841 Znf_RING
IPR036305 RGS_sf
IPR000961 AGC-kinase_C
IPR000719 Prot_kinase_dom
IPR000239 GPCR_kinase
IPR017441 Protein_kinase_ATP_BS
IPR011009 Kinase-like_dom_sf
IPR016137 RGS
IPR000253 FHA_dom
IPR021109 Peptidase_aspartic_dom_sf
IPR001969 Aspartic_peptidase_AS
IPR036179 Ig-like_dom_sf
IPR013783 Ig-like_fold
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR036397 RNaseH_sf
IPR005312 DUF1759
IPR001584 Integrase_cat-core
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR041588 Integrase_H2C2
IPR008737 Peptidase_asp_put
IPR001995 Peptidase_A2_cat
IPR001611 Leu-rich_rpt
IPR032675 LRR_dom_sf
IPR001878 Znf_CCHC
IPR006094 Oxid_FAD_bind_N
IPR016166 FAD-bd_PCMH
IPR016164 FAD-linked_Oxase-like_C
IPR016170 Cytok_DH_C_sf
IPR015213 Cholesterol_OX_subst-bd
IPR036318 FAD-bd_PCMH-like_sf
IPR016169 FAD-bd_PCMH_sub2
IPR016167 FAD-bd_PCMH_sub1
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR000477 RT_dom
IPR019786 Zinc_finger_PHD-type_CS
IPR036875 Znf_CCHC_sf
IPR001841 Znf_RING
IPR036305 RGS_sf
IPR000961 AGC-kinase_C
IPR000719 Prot_kinase_dom
IPR000239 GPCR_kinase
IPR017441 Protein_kinase_ATP_BS
IPR011009 Kinase-like_dom_sf
IPR016137 RGS
IPR000253 FHA_dom
IPR021109 Peptidase_aspartic_dom_sf
IPR001969 Aspartic_peptidase_AS
IPR036179 Ig-like_dom_sf
IPR013783 Ig-like_fold
IPR003599 Ig_sub
IPR007110 Ig-like_dom
SUPFAM
ProteinModelPortal
Q2MGA5
Q9NDM9
Q9NDN0
A0A3S2N5J7
A0A2A4IRQ3
A0A0N1IGH3
+ More
A0A0L7LE26 A0A0L7KQM8 A0A226EFR1 K7JA41 A0A226DGY5 A0A0L7L8X1 A0A226D351 A0A226DH22 A0A226DM06 A0A226EKF8 A0A226EDU9 A0A182G7Z0 W8AJC8 A0A2H1VMV3 A0A182HCB4 A0A226E2P7 A0A1B6LK77 A0A2M4CKZ0 A0A182GL35 W8B882 W8AYL5 A0A2M4CKI9 A0A182G5I1 Q76C95 Q76C94 A0A2M4B9W9 A0A2M4B9D0 A0A182HZZ0 A0A182G158 O02007 A0A1W7R6H4 A0A182H4A2 A0A182HEF9 A0A226DIJ1 A0A0L7LJ73 A0A182H5B8 A0A3S2P6R4 A0A1W7R6M1 A0A2M4CVY8 A0A182GPV7 A0A226DTU1 A0A1W7R6F2 A0A146S4B8 A0A0S7G3I4 A0A2M4BC71 A0A2M4BC51 A0A146PP93 A0A1W7R681 A0A146QCZ7 A0A146S1B6 A0A146Q7X0 A0A0S7G7U6 A0A2M4B8T5 A0A2M4B8I4 A0A0S7FZ14 W4YU11 A0A146SP51 A0A034V9M7 A0A2B4RJC8 A0A1Y1LL65 A0A2G8KYS5 W4Y2R6 A0A0P5QIW7 A0A182GKP7 A0A182GRX9 A0A0P6C5E3 A0A1Y1LN76 A0A0P6AZF3 A0A2G8KVJ2 A0A1W7R6F7 A0A182YRW4 A0A2B4SHN3 A0A1B0CYJ8 A0A1S3QGM9 A0A146S2G4 A0A2B4SX75 A0A146S1A2 A0A146T9S8 A0A085N6I6 W4XIU5 G3NC29 W4XIU4 A0A1A8V6P1 W4XIU3 A0A0S7LK46 W4ZHM9 A0A1S3SW27 A0A146TZK4
A0A0L7LE26 A0A0L7KQM8 A0A226EFR1 K7JA41 A0A226DGY5 A0A0L7L8X1 A0A226D351 A0A226DH22 A0A226DM06 A0A226EKF8 A0A226EDU9 A0A182G7Z0 W8AJC8 A0A2H1VMV3 A0A182HCB4 A0A226E2P7 A0A1B6LK77 A0A2M4CKZ0 A0A182GL35 W8B882 W8AYL5 A0A2M4CKI9 A0A182G5I1 Q76C95 Q76C94 A0A2M4B9W9 A0A2M4B9D0 A0A182HZZ0 A0A182G158 O02007 A0A1W7R6H4 A0A182H4A2 A0A182HEF9 A0A226DIJ1 A0A0L7LJ73 A0A182H5B8 A0A3S2P6R4 A0A1W7R6M1 A0A2M4CVY8 A0A182GPV7 A0A226DTU1 A0A1W7R6F2 A0A146S4B8 A0A0S7G3I4 A0A2M4BC71 A0A2M4BC51 A0A146PP93 A0A1W7R681 A0A146QCZ7 A0A146S1B6 A0A146Q7X0 A0A0S7G7U6 A0A2M4B8T5 A0A2M4B8I4 A0A0S7FZ14 W4YU11 A0A146SP51 A0A034V9M7 A0A2B4RJC8 A0A1Y1LL65 A0A2G8KYS5 W4Y2R6 A0A0P5QIW7 A0A182GKP7 A0A182GRX9 A0A0P6C5E3 A0A1Y1LN76 A0A0P6AZF3 A0A2G8KVJ2 A0A1W7R6F7 A0A182YRW4 A0A2B4SHN3 A0A1B0CYJ8 A0A1S3QGM9 A0A146S2G4 A0A2B4SX75 A0A146S1A2 A0A146T9S8 A0A085N6I6 W4XIU5 G3NC29 W4XIU4 A0A1A8V6P1 W4XIU3 A0A0S7LK46 W4ZHM9 A0A1S3SW27 A0A146TZK4
Ontologies
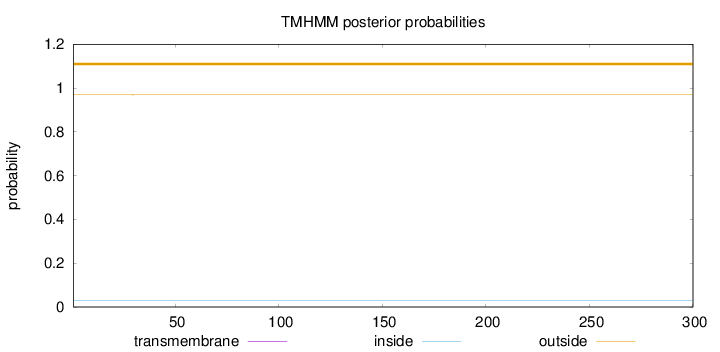
Topology
Length:
300
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00181
Exp number, first 60 AAs:
0.00181
Total prob of N-in:
0.03099
outside
1 - 300
Population Genetic Test Statistics
Pi
353.000644
Theta
169.203029
Tajima's D
3.011953
CLR
1.189128
CSRT
0.980800959952002
Interpretation
Uncertain
