Gene
KWMTBOMO04397
Pre Gene Modal
BGIBMGA005239
Annotation
PREDICTED:_alpha-amylase_4N-like_[Papilio_polytes]
Full name
Alpha-amylase
+ More
Alpha-amylase 4N
Alpha-amylase 4N
Location in the cell
Cytoplasmic Reliability : 1.402 Extracellular Reliability : 2.073
Sequence
CDS
ATGCCTCAGTTGCGATTGCTTCTCGTGTGTTTAATCTTGGGTTTCGTGCTATGCAATCATCATAAAAGGACGAACCAAGATCTCAACCGGTCGACAATTGTGCATCTCTTCGAATGGAAATGGAAGGATATTGCTGATGAATGCGAGAGATTCCTTGCGCCGAAAGGCTTCGGTGGAGTTCAAGTATCTCCACCCTCGGAGAATGTTATCATTCGTTTATCAGACGGTACACGACCTTGGTATGAACGTTACCAAGTAATGTCCTACAAGTTGATCACCAGATCAGGAAATCAAGATGAATTTTTGAACATGACCCGAAGATGTAATGATGTTGGGATAAGAATTTACGCTGACGTGGTAATTAATCATATGACGAGTGACAATAGGGAATCTGTTGGTACAGGGGGTAGTACAGCCAACTATAAAGAGTACAGCTACCCCGCTGTGCCGTACACGAGAGACCACTTCCATCATCCGTCCTGCTCTATTAATAATTATAATGATGCTGCACAGGTAAGGGCCTGCGATTTGGTTGGATTGAAGGACTTGAATCACAACTTGCCGTATGTTCGTGACAAAATCGTTGAATACCTAAACAAGCTTATTGCATTAGGAGTTGCCGGATTCAGAGTGGATGCAGCAAAACATATGTGGCCACACGACCTCAAAGAGATGTACAATAGACTTAATAATCTCAATGTAGATTTTGGCTTCCTACCTAATACAAAACCGTACATTTACCAAGAAGTAATTTATAGAGGGAATGAGCCTATCCAACATACTGAATACACACCCTTTGGTGACGTCACAGAATTTCGGGTCGGATACGAGTTAAAACCAGTTTTTGAGGGTAAAAATCCATTGAAATGGTTGAAGTCCTGGGGCGAGAATTGGAACTTATCACCCAGTGACCGCGTTGTTGTATTCATTGACAATCACGACACGCAGAGGTCGAATGAAGTTTTGACGTACAAAAATGCCAAGTCTTATAAGGCAGCTATAGCGTTTACACTGGCACACCCATATGGCGAACCTCGCGTGATGAGCGGTTATTTCTTCGACAGAACCGACCAAGGTCCACCAAGCGACGGTAACGAGGAAATTTTATCTCCAATCATAAATGATGACGACACCTGCGGCAATGGCTGGGTTTGCGAGCATCGTTGGCGACAGATCTATCAAATGGTCGCATTTAGAAATGCCGTCCGAGATACTAGAGTTGAAAATTGGTGGGACAACGGTCTTAATCAGATAGCTTTCAGTCGAGGAAATAAAGGCTTTATCGCTATTAACGCCGAGGGTCGAGATTTGGATGTTATTTTACAGACTGGTCTTCCATCTGGTACTTATTGCGATGTTATAAGCGGGAAAATTCAAGGCCAAGAATGTTCCGGAAGAAAAATAACAGTTGGCAGTGATGGTCGTGCGCATATTTATGTATCCAAAGACGGGGAGGACATGCATCTTGCCACACATGTTGGGCCGGAGTCATTACTCCAGATCAAGCAGAAGATATGA
Protein
MPQLRLLLVCLILGFVLCNHHKRTNQDLNRSTIVHLFEWKWKDIADECERFLAPKGFGGVQVSPPSENVIIRLSDGTRPWYERYQVMSYKLITRSGNQDEFLNMTRRCNDVGIRIYADVVINHMTSDNRESVGTGGSTANYKEYSYPAVPYTRDHFHHPSCSINNYNDAAQVRACDLVGLKDLNHNLPYVRDKIVEYLNKLIALGVAGFRVDAAKHMWPHDLKEMYNRLNNLNVDFGFLPNTKPYIYQEVIYRGNEPIQHTEYTPFGDVTEFRVGYELKPVFEGKNPLKWLKSWGENWNLSPSDRVVVFIDNHDTQRSNEVLTYKNAKSYKAAIAFTLAHPYGEPRVMSGYFFDRTDQGPPSDGNEEILSPIINDDDTCGNGWVCEHRWRQIYQMVAFRNAVRDTRVENWWDNGLNQIAFSRGNKGFIAINAEGRDLDVILQTGLPSGTYCDVISGKIQGQECSGRKITVGSDGRAHIYVSKDGEDMHLATHVGPESLLQIKQKI
Summary
Catalytic Activity
Endohydrolysis of (1->4)-alpha-D-glucosidic linkages in polysaccharides containing three or more (1->4)-alpha-linked D-glucose units.
Cofactor
Ca(2+)
chloride
chloride
Subunit
Monomer.
Similarity
Belongs to the glycosyl hydrolase 13 family.
Keywords
Calcium
Carbohydrate metabolism
Chloride
Complete proteome
Disulfide bond
Glycosidase
Hydrolase
Metal-binding
Reference proteome
Signal
Feature
chain Alpha-amylase
Uniprot
H9J6U7
A0A3S2LDZ6
A0A0N0PEK4
A0A212FJR5
A0A1I8PGY5
A0A2H1VM91
+ More
A0A212FLR1 A0A0L0CFJ8 A0A2H1X0X9 T1P7P3 A0A2H1WIR2 A0A3G5BII4 A0A1I8MTI4 H9IX93 A0A2A4IUB0 A0A1W4XRV5 A0A1L8EHU5 A0A212FJQ5 Q7YXJ3 A0A0K8W605 Q25590 A0A2A4JNE0 E5LCR9 A0A034VBJ8 Q7YXU5 F6K706 A0A194Q7A8 A0A0K8UF81 A0A034V839 A9XTK5 B1NLD4 A0A182Q521 W8BSY0 W8BYH3 A0A0K8V452 B8Y698 A0A3S2TT63 Q8I9Q2 A0A1B0GIQ4 C7EMF0 H9J6U8 A0A3G5ARJ9 A0A194Q160 A0A034VAD3 A0A1L2FPB8 Q8IA48 A0A182T4P5 Q8MY47 A0A1W4VD39 Q9NKY6 A0A2A4IVZ7 Q8MY50 Q9NKY7 A0A182PEK3 A0A182W2N9 Q8MY55 A0A0A1XMQ5 Q8MY54 A0A1L2FPA6 A0A084WB61 A0A182XVG1 Q8MY53 Q7YXJ4 A0A1I8JUF9 Q24613 Q8IA46 Q9NKZ1 A0A182LXD6 A0A1I8JUE1 Q9NKZ0 Q9NKZ2 Q8N0P9 Q8N0P5 Q9NKY5 O77407 A0A0P8Y7C4 B3MFX1 Q23834 B4KSB4 Q9N651 U5ETD3 Q8N0P8 Q8N0Q8 Q8MM80 Q8MY51 Q8MY52 Q9NKZ3 Q8MY49 Q8N0Q0 Q8N0Q4 Q8MM99 Q9N6Q7 Q8MY48 Q9GRF6 Q8N0Q2 A0A1L2FPA7 A0A182LFL7 Q8MM40 Q7PRB7 A0A182V378
A0A212FLR1 A0A0L0CFJ8 A0A2H1X0X9 T1P7P3 A0A2H1WIR2 A0A3G5BII4 A0A1I8MTI4 H9IX93 A0A2A4IUB0 A0A1W4XRV5 A0A1L8EHU5 A0A212FJQ5 Q7YXJ3 A0A0K8W605 Q25590 A0A2A4JNE0 E5LCR9 A0A034VBJ8 Q7YXU5 F6K706 A0A194Q7A8 A0A0K8UF81 A0A034V839 A9XTK5 B1NLD4 A0A182Q521 W8BSY0 W8BYH3 A0A0K8V452 B8Y698 A0A3S2TT63 Q8I9Q2 A0A1B0GIQ4 C7EMF0 H9J6U8 A0A3G5ARJ9 A0A194Q160 A0A034VAD3 A0A1L2FPB8 Q8IA48 A0A182T4P5 Q8MY47 A0A1W4VD39 Q9NKY6 A0A2A4IVZ7 Q8MY50 Q9NKY7 A0A182PEK3 A0A182W2N9 Q8MY55 A0A0A1XMQ5 Q8MY54 A0A1L2FPA6 A0A084WB61 A0A182XVG1 Q8MY53 Q7YXJ4 A0A1I8JUF9 Q24613 Q8IA46 Q9NKZ1 A0A182LXD6 A0A1I8JUE1 Q9NKZ0 Q9NKZ2 Q8N0P9 Q8N0P5 Q9NKY5 O77407 A0A0P8Y7C4 B3MFX1 Q23834 B4KSB4 Q9N651 U5ETD3 Q8N0P8 Q8N0Q8 Q8MM80 Q8MY51 Q8MY52 Q9NKZ3 Q8MY49 Q8N0Q0 Q8N0Q4 Q8MM99 Q9N6Q7 Q8MY48 Q9GRF6 Q8N0Q2 A0A1L2FPA7 A0A182LFL7 Q8MM40 Q7PRB7 A0A182V378
EC Number
3.2.1.1
Pubmed
EMBL
BABH01028504
RSAL01000030
RVE51685.1
KQ459761
KPJ20139.1
AGBW02008240
+ More
OWR53974.1 ODYU01003330 SOQ41941.1 AGBW02007717 OWR54671.1 JRES01000462 KNC31016.1 ODYU01012544 SOQ58927.1 KA644631 AFP59260.1 ODYU01008873 SOQ52846.1 MK075190 AYV99593.1 BABH01018101 BABH01018102 BABH01018103 BABH01018104 NWSH01006767 PCG63259.1 GFDG01000548 JAV18251.1 OWR53975.1 AY333762 AAP97394.1 GDHF01005753 JAI46561.1 U04223 AAA03715.1 NWSH01001047 PCG72932.1 HQ424576 ADP89000.1 GAKP01019473 GAKP01019470 JAC39482.1 AY330289 AAP92665.1 HM357843 AEA76309.1 KQ459580 KPI99290.1 GDHF01027086 JAI25228.1 GAKP01019466 JAC39486.1 EF672102 ABV24963.1 EF600049 EU325552 ABU98614.1 ACB54942.1 AXCN02001626 GAMC01004278 JAC02278.1 GAMC01008204 JAB98351.1 GDHF01018640 JAI33674.1 FJ489868 ACL14798.1 RSAL01000004 RVE54528.1 AF467104 AAO17923.2 AJWK01015266 AJWK01015267 GQ274006 ACT64133.1 BABH01028500 BABH01028501 BABH01028502 MG941008 AYV89929.1 KPI99291.1 GAKP01019468 JAC39484.1 KX398855 ANZ79224.1 AF146757 AAO13691.1 AB078773 BAC06339.1 AB035066 BAA95440.1 NWSH01006246 PCG63558.1 AB078770 BAC06336.1 AB035065 BAA95439.1 AB078765 BAC06331.1 GBXI01001663 JAD12629.1 AB078766 BAC06332.1 KX398852 ANZ79221.1 ATLV01022314 KE525331 KFB47455.1 AB078767 BAC06333.1 AY333761 AAP97393.1 X76241 CAA53820.1 AF280891 AAO13754.1 AB035061 BAA95435.1 AXCM01000750 AB035062 BAA95436.1 AB035060 AB077431 BAA95434.1 BAB91544.1 AB077432 BAB91545.1 AB077436 BAB91549.1 AB035069 BAA95443.1 U53699 AAC35243.1 CH902619 KPU77312.1 EDV35653.1 U53477 U31121 CH933808 EDW08396.1 AB035057 AB035058 BAA95432.1 GANO01001973 JAB57898.1 AB077433 BAB91546.1 AB077391 BAB91504.1 AB077412 AB077413 AB077417 AB077423 AB077424 BAB91525.1 BAB91526.1 BAB91530.1 BAB91536.1 BAB91537.1 AB078769 BAC06335.1 AB078768 BAC06334.1 AB035059 BAA95433.1 AB078771 BAC06337.1 AB077430 BAB91543.1 AB077404 BAB91517.1 AB077388 AB077395 AB077406 AB077407 BAB91501.1 BAB91508.1 BAB91519.1 BAB91520.1 AB035067 AB035068 BAA95442.1 AB078772 BAC06338.1 U53478 AAB69330.1 AB077426 BAB91539.1 KX398853 ANZ79222.1 AB077409 AB077410 AB077411 AB077415 AB077421 AB077422 AB077425 AB077428 AB077429 BAB91522.1 BAB91523.1 BAB91524.1 BAB91528.1 BAB91534.1 BAB91535.1 BAB91538.1 BAB91541.1 BAB91542.1 AAAB01008859 EAA07513.6
OWR53974.1 ODYU01003330 SOQ41941.1 AGBW02007717 OWR54671.1 JRES01000462 KNC31016.1 ODYU01012544 SOQ58927.1 KA644631 AFP59260.1 ODYU01008873 SOQ52846.1 MK075190 AYV99593.1 BABH01018101 BABH01018102 BABH01018103 BABH01018104 NWSH01006767 PCG63259.1 GFDG01000548 JAV18251.1 OWR53975.1 AY333762 AAP97394.1 GDHF01005753 JAI46561.1 U04223 AAA03715.1 NWSH01001047 PCG72932.1 HQ424576 ADP89000.1 GAKP01019473 GAKP01019470 JAC39482.1 AY330289 AAP92665.1 HM357843 AEA76309.1 KQ459580 KPI99290.1 GDHF01027086 JAI25228.1 GAKP01019466 JAC39486.1 EF672102 ABV24963.1 EF600049 EU325552 ABU98614.1 ACB54942.1 AXCN02001626 GAMC01004278 JAC02278.1 GAMC01008204 JAB98351.1 GDHF01018640 JAI33674.1 FJ489868 ACL14798.1 RSAL01000004 RVE54528.1 AF467104 AAO17923.2 AJWK01015266 AJWK01015267 GQ274006 ACT64133.1 BABH01028500 BABH01028501 BABH01028502 MG941008 AYV89929.1 KPI99291.1 GAKP01019468 JAC39484.1 KX398855 ANZ79224.1 AF146757 AAO13691.1 AB078773 BAC06339.1 AB035066 BAA95440.1 NWSH01006246 PCG63558.1 AB078770 BAC06336.1 AB035065 BAA95439.1 AB078765 BAC06331.1 GBXI01001663 JAD12629.1 AB078766 BAC06332.1 KX398852 ANZ79221.1 ATLV01022314 KE525331 KFB47455.1 AB078767 BAC06333.1 AY333761 AAP97393.1 X76241 CAA53820.1 AF280891 AAO13754.1 AB035061 BAA95435.1 AXCM01000750 AB035062 BAA95436.1 AB035060 AB077431 BAA95434.1 BAB91544.1 AB077432 BAB91545.1 AB077436 BAB91549.1 AB035069 BAA95443.1 U53699 AAC35243.1 CH902619 KPU77312.1 EDV35653.1 U53477 U31121 CH933808 EDW08396.1 AB035057 AB035058 BAA95432.1 GANO01001973 JAB57898.1 AB077433 BAB91546.1 AB077391 BAB91504.1 AB077412 AB077413 AB077417 AB077423 AB077424 BAB91525.1 BAB91526.1 BAB91530.1 BAB91536.1 BAB91537.1 AB078769 BAC06335.1 AB078768 BAC06334.1 AB035059 BAA95433.1 AB078771 BAC06337.1 AB077430 BAB91543.1 AB077404 BAB91517.1 AB077388 AB077395 AB077406 AB077407 BAB91501.1 BAB91508.1 BAB91519.1 BAB91520.1 AB035067 AB035068 BAA95442.1 AB078772 BAC06338.1 U53478 AAB69330.1 AB077426 BAB91539.1 KX398853 ANZ79222.1 AB077409 AB077410 AB077411 AB077415 AB077421 AB077422 AB077425 AB077428 AB077429 BAB91522.1 BAB91523.1 BAB91524.1 BAB91528.1 BAB91534.1 BAB91535.1 BAB91538.1 BAB91541.1 BAB91542.1 AAAB01008859 EAA07513.6
Proteomes
UP000005204
UP000283053
UP000053240
UP000007151
UP000095300
UP000037069
+ More
UP000095301 UP000218220 UP000192223 UP000053268 UP000075886 UP000092461 UP000075901 UP000192221 UP000075885 UP000075920 UP000030765 UP000076408 UP000075900 UP000075883 UP000007801 UP000009192 UP000075882 UP000007062 UP000075903
UP000095301 UP000218220 UP000192223 UP000053268 UP000075886 UP000092461 UP000075901 UP000192221 UP000075885 UP000075920 UP000030765 UP000076408 UP000075900 UP000075883 UP000007801 UP000009192 UP000075882 UP000007062 UP000075903
Interpro
IPR006047
Glyco_hydro_13_cat_dom
+ More
IPR017853 Glycoside_hydrolase_SF
IPR006046 Alpha_amylase
IPR031319 A-amylase_C
IPR006048 A-amylase/branching_C
IPR013780 Glyco_hydro_b
IPR015421 PyrdxlP-dep_Trfase_major
IPR015424 PyrdxlP-dep_Trfase
IPR004839 Aminotransferase_I/II
IPR001917 Aminotrans_II_pyridoxalP_BS
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR017853 Glycoside_hydrolase_SF
IPR006046 Alpha_amylase
IPR031319 A-amylase_C
IPR006048 A-amylase/branching_C
IPR013780 Glyco_hydro_b
IPR015421 PyrdxlP-dep_Trfase_major
IPR015424 PyrdxlP-dep_Trfase
IPR004839 Aminotransferase_I/II
IPR001917 Aminotrans_II_pyridoxalP_BS
IPR015422 PyrdxlP-dep_Trfase_dom1
Gene 3D
ProteinModelPortal
H9J6U7
A0A3S2LDZ6
A0A0N0PEK4
A0A212FJR5
A0A1I8PGY5
A0A2H1VM91
+ More
A0A212FLR1 A0A0L0CFJ8 A0A2H1X0X9 T1P7P3 A0A2H1WIR2 A0A3G5BII4 A0A1I8MTI4 H9IX93 A0A2A4IUB0 A0A1W4XRV5 A0A1L8EHU5 A0A212FJQ5 Q7YXJ3 A0A0K8W605 Q25590 A0A2A4JNE0 E5LCR9 A0A034VBJ8 Q7YXU5 F6K706 A0A194Q7A8 A0A0K8UF81 A0A034V839 A9XTK5 B1NLD4 A0A182Q521 W8BSY0 W8BYH3 A0A0K8V452 B8Y698 A0A3S2TT63 Q8I9Q2 A0A1B0GIQ4 C7EMF0 H9J6U8 A0A3G5ARJ9 A0A194Q160 A0A034VAD3 A0A1L2FPB8 Q8IA48 A0A182T4P5 Q8MY47 A0A1W4VD39 Q9NKY6 A0A2A4IVZ7 Q8MY50 Q9NKY7 A0A182PEK3 A0A182W2N9 Q8MY55 A0A0A1XMQ5 Q8MY54 A0A1L2FPA6 A0A084WB61 A0A182XVG1 Q8MY53 Q7YXJ4 A0A1I8JUF9 Q24613 Q8IA46 Q9NKZ1 A0A182LXD6 A0A1I8JUE1 Q9NKZ0 Q9NKZ2 Q8N0P9 Q8N0P5 Q9NKY5 O77407 A0A0P8Y7C4 B3MFX1 Q23834 B4KSB4 Q9N651 U5ETD3 Q8N0P8 Q8N0Q8 Q8MM80 Q8MY51 Q8MY52 Q9NKZ3 Q8MY49 Q8N0Q0 Q8N0Q4 Q8MM99 Q9N6Q7 Q8MY48 Q9GRF6 Q8N0Q2 A0A1L2FPA7 A0A182LFL7 Q8MM40 Q7PRB7 A0A182V378
A0A212FLR1 A0A0L0CFJ8 A0A2H1X0X9 T1P7P3 A0A2H1WIR2 A0A3G5BII4 A0A1I8MTI4 H9IX93 A0A2A4IUB0 A0A1W4XRV5 A0A1L8EHU5 A0A212FJQ5 Q7YXJ3 A0A0K8W605 Q25590 A0A2A4JNE0 E5LCR9 A0A034VBJ8 Q7YXU5 F6K706 A0A194Q7A8 A0A0K8UF81 A0A034V839 A9XTK5 B1NLD4 A0A182Q521 W8BSY0 W8BYH3 A0A0K8V452 B8Y698 A0A3S2TT63 Q8I9Q2 A0A1B0GIQ4 C7EMF0 H9J6U8 A0A3G5ARJ9 A0A194Q160 A0A034VAD3 A0A1L2FPB8 Q8IA48 A0A182T4P5 Q8MY47 A0A1W4VD39 Q9NKY6 A0A2A4IVZ7 Q8MY50 Q9NKY7 A0A182PEK3 A0A182W2N9 Q8MY55 A0A0A1XMQ5 Q8MY54 A0A1L2FPA6 A0A084WB61 A0A182XVG1 Q8MY53 Q7YXJ4 A0A1I8JUF9 Q24613 Q8IA46 Q9NKZ1 A0A182LXD6 A0A1I8JUE1 Q9NKZ0 Q9NKZ2 Q8N0P9 Q8N0P5 Q9NKY5 O77407 A0A0P8Y7C4 B3MFX1 Q23834 B4KSB4 Q9N651 U5ETD3 Q8N0P8 Q8N0Q8 Q8MM80 Q8MY51 Q8MY52 Q9NKZ3 Q8MY49 Q8N0Q0 Q8N0Q4 Q8MM99 Q9N6Q7 Q8MY48 Q9GRF6 Q8N0Q2 A0A1L2FPA7 A0A182LFL7 Q8MM40 Q7PRB7 A0A182V378
PDB
1VIW
E-value=7.36993e-157,
Score=1422
Ontologies
PATHWAY
GO
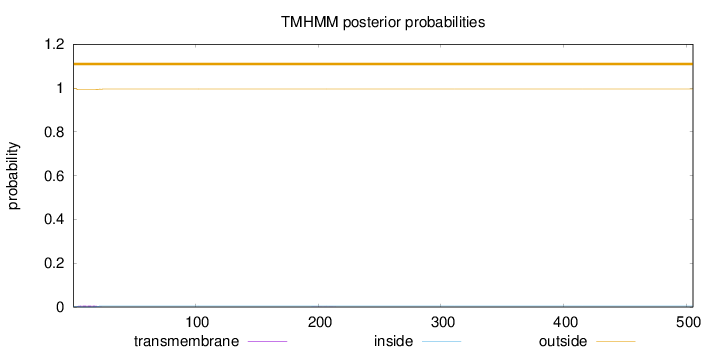
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.967376

Length:
505
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11163
Exp number, first 60 AAs:
0.10587
Total prob of N-in:
0.00273
outside
1 - 505
Population Genetic Test Statistics
Pi
240.824248
Theta
168.302705
Tajima's D
1.385034
CLR
0.258557
CSRT
0.75741212939353
Interpretation
Uncertain
