Gene
KWMTBOMO04394
Annotation
endonuclease_and_reverse_transcriptase-like_protein_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.543 Nuclear Reliability : 1.221
Sequence
CDS
ATGGTGCAAGAGAGACAATCAAAACTTGTGTCTGAAGTGGAGAACTCTCTGGGGCGAGTCTCCGAATGGGGTGAATTGAACTTGGTTCAATTCAACCCGATAAAGACACAAGTTTGCGCGTTCACTGCGAAGAAGGACCCCTCTGTCATGGCGCCGCAATTCCAAGGAGTATCCCTGCAACCTTCCGAGAGTATCGGGATACTTGGGGTCGACATTTCGAGCGATGTCCAATTTCGGAGTCATTTGGAAGGCAAAGCCAAGTTGGCGTCCAAAATGCTGGGAGTCCTCAACAGAGCCAAGCGGTACTTGACACCTGGACAAAGACTTTTGCTCTATAAAGCACAAGTCCGGCCTCGCGTGGAGTACTGCTCCCATCTCTGGGCCGGGGCTCCCAAGTACCAGCTTCTTCCATTTGACTCCATATAG
Protein
MVQERQSKLVSEVENSLGRVSEWGELNLVQFNPIKTQVCAFTAKKDPSVMAPQFQGVSLQPSESIGILGVDISSDVQFRSHLEGKAKLASKMLGVLNRAKRYLTPGQRLLLYKAQVRPRVEYCSHLWAGAPKYQLLPFDSI
Summary
Cofactor
pyridoxal 5'-phosphate
Uniprot
Q6UV17
Q5KTM5
A0A3S2KYN2
B9WPT7
A0A3S2LP03
A0A3S2NMX8
+ More
A0A3S2LQP1 A0A3S2L424 A0A3S2NTH0 A0A3S2NHP4 A0A3S2NN41 A0A2H1VFZ3 A0A3S2M4N8 A0A2H1WCG9 A0A3S2NMS5 A0A3S2P9Q3 A0A3S2TCJ2 J9GT67 A0A0P4VXU0 A0A2H1V9A8 A0A3D5S1K1 A0A2H1V1V5 T2M6M7 A0A3S2PCY7 A0A1Y1S3P2 A0A3S2TFH4 A0A0K8RPQ6 A0A2H9T325 T2M4I6 V5H8T0 A0A147BI04 A0A1E1XGQ7 A0A131XV56 A0A3Q0IWM1 B7P320 A0A147BKN8 A0A0P4VPK5 A0A2S2PKE0 A0A0P4VVV8 A0A147BBR8 A0A090XE26 A0A131Y4I8 A0A1E1XJ46 A0A147BU27 A0A3S2P593 A0A090X7P7 A0A1B6HRT6 A0A131XRE4 A0A147BJQ0 A0A3P9H8U4 A0A1G4M5I2 A0A3B3I2N9 A0A3P9JA17 A0A3P9IMI8 A0A3P9JPR7 A0A3B3HJ43 A0A3P9JG11 A0A3P9IP68 A0A3P9IN24 A0A3P9IMZ6 A0A131XU84 A0A3P9H763 A0A1E1XMS7 A0A1B6JHC3 A0A293LGC3 A0A3B3HUG5 A0A3P9L6L3 A0A3P9H895 A0A3B3HPJ3 A0A3P9H463 A0A3P9HWJ2 A0A3P9I7G8 A0A3P9IY49 A0A3P9IRG5 A0A3P9KUJ9 A0A3P9HBD7 A0A3P9H9P0 A0A147BQ97 A0A3P9JEY5 A0A3B3HGP3 A0A3B3IP08 A0A182GLR7 A0A3M6TMT5 A0A3B3HSG8 A0A3B3H5R2 A0A3B3I4F1 A0A3B3I465 A0A3P9JUY5 A0A3B3HFB5 A0A3B3I6F0 A0A3B3HAL5 A0A1E1XNL1 A0A3B3H5K9
A0A3S2LQP1 A0A3S2L424 A0A3S2NTH0 A0A3S2NHP4 A0A3S2NN41 A0A2H1VFZ3 A0A3S2M4N8 A0A2H1WCG9 A0A3S2NMS5 A0A3S2P9Q3 A0A3S2TCJ2 J9GT67 A0A0P4VXU0 A0A2H1V9A8 A0A3D5S1K1 A0A2H1V1V5 T2M6M7 A0A3S2PCY7 A0A1Y1S3P2 A0A3S2TFH4 A0A0K8RPQ6 A0A2H9T325 T2M4I6 V5H8T0 A0A147BI04 A0A1E1XGQ7 A0A131XV56 A0A3Q0IWM1 B7P320 A0A147BKN8 A0A0P4VPK5 A0A2S2PKE0 A0A0P4VVV8 A0A147BBR8 A0A090XE26 A0A131Y4I8 A0A1E1XJ46 A0A147BU27 A0A3S2P593 A0A090X7P7 A0A1B6HRT6 A0A131XRE4 A0A147BJQ0 A0A3P9H8U4 A0A1G4M5I2 A0A3B3I2N9 A0A3P9JA17 A0A3P9IMI8 A0A3P9JPR7 A0A3B3HJ43 A0A3P9JG11 A0A3P9IP68 A0A3P9IN24 A0A3P9IMZ6 A0A131XU84 A0A3P9H763 A0A1E1XMS7 A0A1B6JHC3 A0A293LGC3 A0A3B3HUG5 A0A3P9L6L3 A0A3P9H895 A0A3B3HPJ3 A0A3P9H463 A0A3P9HWJ2 A0A3P9I7G8 A0A3P9IY49 A0A3P9IRG5 A0A3P9KUJ9 A0A3P9HBD7 A0A3P9H9P0 A0A147BQ97 A0A3P9JEY5 A0A3B3HGP3 A0A3B3IP08 A0A182GLR7 A0A3M6TMT5 A0A3B3HSG8 A0A3B3H5R2 A0A3B3I4F1 A0A3B3I465 A0A3P9JUY5 A0A3B3HFB5 A0A3B3I6F0 A0A3B3HAL5 A0A1E1XNL1 A0A3B3H5K9
Pubmed
EMBL
AY359886
AAQ57129.1
AB126052
BAD86652.1
RSAL01000487
RVE41561.1
+ More
FM995623 CAX36787.1 RSAL01002067 RVE40593.1 RSAL01000002 RVE55041.1 RVE55094.1 RSAL01000422 RVE41792.1 RSAL01000171 RVE45150.1 RSAL01000090 RVE48098.1 RSAL01003136 RVE40332.1 RSAL01000028 RVE51880.1 ODYU01002162 SOQ39302.1 RSAL01000035 RVE51317.1 ODYU01007423 SOQ50184.1 RVE54942.1 RSAL01000164 RVE45343.1 RSAL01000380 RVE42061.1 AMCI01002135 EJX03495.1 GDRN01105059 JAI57791.1 ODYU01001348 SOQ37407.1 DPOC01000269 HCX22453.1 ODYU01000310 SOQ34825.1 HAAD01001474 CDG67706.1 RSAL01000095 RVE47834.1 LWDP01000391 ORD92879.1 RSAL01000193 RVE44603.1 GADI01001229 JAA72579.1 NSIT01000489 PJE77589.1 HAAD01000593 CDG66825.1 GANP01010944 JAB73524.1 GEGO01005479 JAR89925.1 GFAC01000919 JAT98269.1 GEFM01006250 JAP69546.1 ABJB010171673 DS626007 EEC00992.1 GEGO01004056 JAR91348.1 GDRN01111278 JAI56803.1 GGMR01016717 MBY29336.1 GDRN01111277 JAI56804.1 GEGO01007187 JAR88217.1 GBIH01002577 JAC92133.1 GEFM01002394 JAP73402.1 GFAA01004201 JAT99233.1 GEGO01001118 JAR94286.1 RSAL01000369 RVE42156.1 GBIH01002574 JAC92136.1 GECU01030313 JAS77393.1 GEFM01005882 JAP69914.1 GEGO01004433 JAR90971.1 FLMD02000280 SCV66640.1 GEFM01005904 JAP69892.1 GFAA01002794 JAU00641.1 GECU01009092 JAS98614.1 GFWV01002824 MAA27554.1 GEGO01002473 JAR92931.1 JXUM01073786 KQ562795 KXJ75115.1 RCHS01003332 RMX42564.1 GFAA01002514 JAU00921.1
FM995623 CAX36787.1 RSAL01002067 RVE40593.1 RSAL01000002 RVE55041.1 RVE55094.1 RSAL01000422 RVE41792.1 RSAL01000171 RVE45150.1 RSAL01000090 RVE48098.1 RSAL01003136 RVE40332.1 RSAL01000028 RVE51880.1 ODYU01002162 SOQ39302.1 RSAL01000035 RVE51317.1 ODYU01007423 SOQ50184.1 RVE54942.1 RSAL01000164 RVE45343.1 RSAL01000380 RVE42061.1 AMCI01002135 EJX03495.1 GDRN01105059 JAI57791.1 ODYU01001348 SOQ37407.1 DPOC01000269 HCX22453.1 ODYU01000310 SOQ34825.1 HAAD01001474 CDG67706.1 RSAL01000095 RVE47834.1 LWDP01000391 ORD92879.1 RSAL01000193 RVE44603.1 GADI01001229 JAA72579.1 NSIT01000489 PJE77589.1 HAAD01000593 CDG66825.1 GANP01010944 JAB73524.1 GEGO01005479 JAR89925.1 GFAC01000919 JAT98269.1 GEFM01006250 JAP69546.1 ABJB010171673 DS626007 EEC00992.1 GEGO01004056 JAR91348.1 GDRN01111278 JAI56803.1 GGMR01016717 MBY29336.1 GDRN01111277 JAI56804.1 GEGO01007187 JAR88217.1 GBIH01002577 JAC92133.1 GEFM01002394 JAP73402.1 GFAA01004201 JAT99233.1 GEGO01001118 JAR94286.1 RSAL01000369 RVE42156.1 GBIH01002574 JAC92136.1 GECU01030313 JAS77393.1 GEFM01005882 JAP69914.1 GEGO01004433 JAR90971.1 FLMD02000280 SCV66640.1 GEFM01005904 JAP69892.1 GFAA01002794 JAU00641.1 GECU01009092 JAS98614.1 GFWV01002824 MAA27554.1 GEGO01002473 JAR92931.1 JXUM01073786 KQ562795 KXJ75115.1 RCHS01003332 RMX42564.1 GFAA01002514 JAU00921.1
Proteomes
Pfam
Interpro
IPR000477
RT_dom
+ More
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR034653 SPF45_RRM
IPR003954 RRM_dom_euk
IPR000467 G_patch_dom
IPR040052 RBM17
IPR035979 RBD_domain_sf
IPR015424 PyrdxlP-dep_Trfase
IPR002129 PyrdxlP-dep_de-COase
IPR021115 Pyridoxal-P_BS
IPR015421 PyrdxlP-dep_Trfase_major
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR010977 Aromatic_deC
IPR007023 Ribosom_reg
IPR019442 DUF2428_death-receptor-like
IPR016190 Transl_init_fac_IF2/IF5_Zn-bd
IPR002735 Transl_init_fac_IF2/IF5
IPR016189 Transl_init_fac_IF2/IF5_N
IPR039779 RFX-like
IPR018957 Znf_C3HC4_RING-type
IPR036987 SRA-YDG_sf
IPR013083 Znf_RING/FYVE/PHD
IPR017907 Znf_RING_CS
IPR015947 PUA-like_sf
IPR003105 SRA_YDG
IPR001841 Znf_RING
IPR036322 WD40_repeat_dom_sf
IPR040382 NOL10/Enp2
IPR012580 NUC153
IPR001680 WD40_repeat
IPR016159 Cullin_repeat-like_dom_sf
IPR004140 Exo70
IPR000738 WHEP-TRS_dom
IPR009068 S15_NS1_RNA-bd
IPR030223 Dispatched
IPR003392 Ptc/Disp
IPR000731 SSD
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR034653 SPF45_RRM
IPR003954 RRM_dom_euk
IPR000467 G_patch_dom
IPR040052 RBM17
IPR035979 RBD_domain_sf
IPR015424 PyrdxlP-dep_Trfase
IPR002129 PyrdxlP-dep_de-COase
IPR021115 Pyridoxal-P_BS
IPR015421 PyrdxlP-dep_Trfase_major
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR010977 Aromatic_deC
IPR007023 Ribosom_reg
IPR019442 DUF2428_death-receptor-like
IPR016190 Transl_init_fac_IF2/IF5_Zn-bd
IPR002735 Transl_init_fac_IF2/IF5
IPR016189 Transl_init_fac_IF2/IF5_N
IPR039779 RFX-like
IPR018957 Znf_C3HC4_RING-type
IPR036987 SRA-YDG_sf
IPR013083 Znf_RING/FYVE/PHD
IPR017907 Znf_RING_CS
IPR015947 PUA-like_sf
IPR003105 SRA_YDG
IPR001841 Znf_RING
IPR036322 WD40_repeat_dom_sf
IPR040382 NOL10/Enp2
IPR012580 NUC153
IPR001680 WD40_repeat
IPR016159 Cullin_repeat-like_dom_sf
IPR004140 Exo70
IPR000738 WHEP-TRS_dom
IPR009068 S15_NS1_RNA-bd
IPR030223 Dispatched
IPR003392 Ptc/Disp
IPR000731 SSD
SUPFAM
ProteinModelPortal
Q6UV17
Q5KTM5
A0A3S2KYN2
B9WPT7
A0A3S2LP03
A0A3S2NMX8
+ More
A0A3S2LQP1 A0A3S2L424 A0A3S2NTH0 A0A3S2NHP4 A0A3S2NN41 A0A2H1VFZ3 A0A3S2M4N8 A0A2H1WCG9 A0A3S2NMS5 A0A3S2P9Q3 A0A3S2TCJ2 J9GT67 A0A0P4VXU0 A0A2H1V9A8 A0A3D5S1K1 A0A2H1V1V5 T2M6M7 A0A3S2PCY7 A0A1Y1S3P2 A0A3S2TFH4 A0A0K8RPQ6 A0A2H9T325 T2M4I6 V5H8T0 A0A147BI04 A0A1E1XGQ7 A0A131XV56 A0A3Q0IWM1 B7P320 A0A147BKN8 A0A0P4VPK5 A0A2S2PKE0 A0A0P4VVV8 A0A147BBR8 A0A090XE26 A0A131Y4I8 A0A1E1XJ46 A0A147BU27 A0A3S2P593 A0A090X7P7 A0A1B6HRT6 A0A131XRE4 A0A147BJQ0 A0A3P9H8U4 A0A1G4M5I2 A0A3B3I2N9 A0A3P9JA17 A0A3P9IMI8 A0A3P9JPR7 A0A3B3HJ43 A0A3P9JG11 A0A3P9IP68 A0A3P9IN24 A0A3P9IMZ6 A0A131XU84 A0A3P9H763 A0A1E1XMS7 A0A1B6JHC3 A0A293LGC3 A0A3B3HUG5 A0A3P9L6L3 A0A3P9H895 A0A3B3HPJ3 A0A3P9H463 A0A3P9HWJ2 A0A3P9I7G8 A0A3P9IY49 A0A3P9IRG5 A0A3P9KUJ9 A0A3P9HBD7 A0A3P9H9P0 A0A147BQ97 A0A3P9JEY5 A0A3B3HGP3 A0A3B3IP08 A0A182GLR7 A0A3M6TMT5 A0A3B3HSG8 A0A3B3H5R2 A0A3B3I4F1 A0A3B3I465 A0A3P9JUY5 A0A3B3HFB5 A0A3B3I6F0 A0A3B3HAL5 A0A1E1XNL1 A0A3B3H5K9
A0A3S2LQP1 A0A3S2L424 A0A3S2NTH0 A0A3S2NHP4 A0A3S2NN41 A0A2H1VFZ3 A0A3S2M4N8 A0A2H1WCG9 A0A3S2NMS5 A0A3S2P9Q3 A0A3S2TCJ2 J9GT67 A0A0P4VXU0 A0A2H1V9A8 A0A3D5S1K1 A0A2H1V1V5 T2M6M7 A0A3S2PCY7 A0A1Y1S3P2 A0A3S2TFH4 A0A0K8RPQ6 A0A2H9T325 T2M4I6 V5H8T0 A0A147BI04 A0A1E1XGQ7 A0A131XV56 A0A3Q0IWM1 B7P320 A0A147BKN8 A0A0P4VPK5 A0A2S2PKE0 A0A0P4VVV8 A0A147BBR8 A0A090XE26 A0A131Y4I8 A0A1E1XJ46 A0A147BU27 A0A3S2P593 A0A090X7P7 A0A1B6HRT6 A0A131XRE4 A0A147BJQ0 A0A3P9H8U4 A0A1G4M5I2 A0A3B3I2N9 A0A3P9JA17 A0A3P9IMI8 A0A3P9JPR7 A0A3B3HJ43 A0A3P9JG11 A0A3P9IP68 A0A3P9IN24 A0A3P9IMZ6 A0A131XU84 A0A3P9H763 A0A1E1XMS7 A0A1B6JHC3 A0A293LGC3 A0A3B3HUG5 A0A3P9L6L3 A0A3P9H895 A0A3B3HPJ3 A0A3P9H463 A0A3P9HWJ2 A0A3P9I7G8 A0A3P9IY49 A0A3P9IRG5 A0A3P9KUJ9 A0A3P9HBD7 A0A3P9H9P0 A0A147BQ97 A0A3P9JEY5 A0A3B3HGP3 A0A3B3IP08 A0A182GLR7 A0A3M6TMT5 A0A3B3HSG8 A0A3B3H5R2 A0A3B3I4F1 A0A3B3I465 A0A3P9JUY5 A0A3B3HFB5 A0A3B3I6F0 A0A3B3HAL5 A0A1E1XNL1 A0A3B3H5K9
Ontologies
Topology
Subcellular location
Nucleus
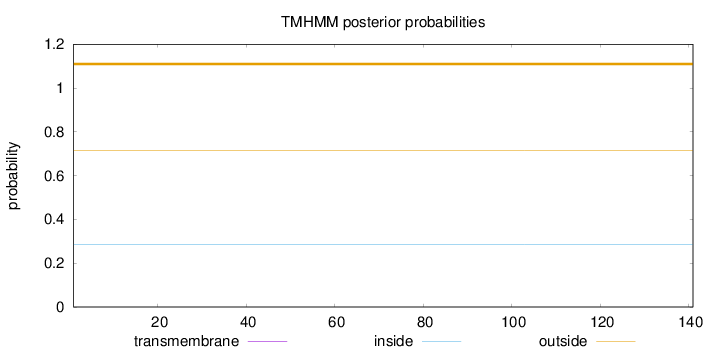
Length:
141
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00073
Exp number, first 60 AAs:
8e-05
Total prob of N-in:
0.28406
outside
1 - 141
Population Genetic Test Statistics
Pi
260.559076
Theta
143.204013
Tajima's D
2.57548
CLR
0.492911
CSRT
0.950502474876256
Interpretation
Uncertain
