Gene
KWMTBOMO04388
Pre Gene Modal
BGIBMGA005281
Annotation
PREDICTED:_serine-enriched_protein_isoform_X3_[Bombyx_mori]
Full name
Alpha-amylase
+ More
Serine-enriched protein
Serine-enriched protein
Location in the cell
Nuclear Reliability : 2.461
Sequence
CDS
ATGAGAAGTGGGGCGTTAGCGGCTATGGAGGCGCAGCCTGACCTCAGCACCTTCGAGAACAAGAGCGGGCTGGCGGAGGACATGAAGTTCCTGGCGTCGATGCCTGAGCTCTGTGACGTCACCTTCCTGGTGGGAGACACCAGGGAGCCCGTTTGTGCTGTGAAAGCGGTCCTGGCGGCAAGGAGCAGAGTCTTCCAAAAAATGCTATATCAAGCTCCAAGTCCTCAGAGAAAGAAGGAACCAGGACCAAGAGAGAACAAGCTAAGATTATTCTTGAAACGTTCCTCGGAGCCTCTTCTGAATCTACAAAATGCAGCACAGCAGCCTTCTGCTCAGCAACATCAGACCTTAATAATCGAAGAATTTGAACCGGACGTTTTTCGTCAACTAATCGAGTATATCCATACCGGATGTGTTACTCTTCAACCAAGAACTCTTCTGGGAGTTATGAATGCGGCTGATTACTACGGCTTGGATGAGCTGCGACGAGCTTGCGCTGGTTTCGTACAGTGCTGTATCACCGTGGATACTGTATGCGCATTACTAGCTTCGGCCGAGCGTTACATACAATATAAGTGCACGAAGTCGCTGGTCCAGAAGGTTCTAGAATTCGTCGATGAGCATGGCAATGAGGTATTAAACTTGGGATCCTTCACGCTTCTTCCTCAACATGTTGTCAGATTGATCCTAGCAAGAGATGAACTCCGAGCGGATGAATTCACCAAATTTCAGGCAGCACTTATGTGGGGCAAAAAATTTTGCGACACGAACCAAAACACATCATTGAAAGACGTCATTGGGAATTTCTTGGAATACATACAGTTCCATAGAATTCCAGCGAATGTTCTCATGAGGGAAGTGCATCCGCTGGGCTTGGTACCGTATTCCATTATCATGAACGCTTTGGCGTATCAGGCAGACCCCGCTAGCGTAGATCCAGGGAAACTGTCTCCAGCGAGAATTCGACGAGCCGGCCGGTCGATGTCTGTGCAGTCGTCAATGGATCCCTACGGGTCCAACACCACCCTCTCCTCCACCGGCTCGAGCGATGTTCCGTCTTCAGACTCTCGGCACAACTAA
Protein
MRSGALAAMEAQPDLSTFENKSGLAEDMKFLASMPELCDVTFLVGDTREPVCAVKAVLAARSRVFQKMLYQAPSPQRKKEPGPRENKLRLFLKRSSEPLLNLQNAAQQPSAQQHQTLIIEEFEPDVFRQLIEYIHTGCVTLQPRTLLGVMNAADYYGLDELRRACAGFVQCCITVDTVCALLASAERYIQYKCTKSLVQKVLEFVDEHGNEVLNLGSFTLLPQHVVRLILARDELRADEFTKFQAALMWGKKFCDTNQNTSLKDVIGNFLEYIQFHRIPANVLMREVHPLGLVPYSIIMNALAYQADPASVDPGKLSPARIRRAGRSMSVQSSMDPYGSNTTLSSTGSSDVPSSDSRHN
Summary
Similarity
Belongs to the glycosyl hydrolase 13 family.
Keywords
Complete proteome
Reference proteome
Feature
chain Alpha-amylase
Uniprot
A0A2H1W2A6
A0A2A4J9C8
A0A212ELJ8
A0A0N1IQ65
A0A182R5H0
A0A182VLN3
+ More
A0A182XD91 A0A182K8H4 A0A182PBN9 A0A182M4J2 A0A182NPH1 A0A182KSR9 A0A182Y1V7 A0A182QS62 A0A182GTH1 A0A084WLP8 E0VVS0 W5JQS1 A0A1Y1NDQ5 A0A182G5E8 A0A067RLF1 A0A182J907 A0A182HPZ9 A0A0J7L4B0 A0A1W4X3G2 A0A139WCU8 A0A336MNK9 A0A194Q1U0 A0A336MLJ2 A0A1W4XE57 E2BN74 Q7PV79 Q17M62 A0A1B0DHR1 A0A1I8PPP4 A0A151J7H1 A0A195D726 A0A1I8NIA6 K7IWI8 A0A0P9BZW6 A0A158NFL1 A0A0Q9XA06 A0A0R3NQU5 B0WEI5 A0A2A3EIW1 A0A0J9REF5 A0A0B4KFS9 V9I9S3 V9I9A5 A0A0Q5VLF4 A0A088AEG7 B4JWN0 A0A182FUE2 A0A1I8PPR4 A0A0L0BW10 A0A1A9X2G5 A0A182W989 A0A195AY50 A0A0Q9XAJ0 A0A0R3NQ29 A0A026WS64 A0A1A9XXV3 A0A0B4KEX5 A0A0R1DSS3 A0A0J9RE05 A0A1W4UZH5 A0A0Q5VM06 A0A1B6D7J1 A0A3L8DD70 A0A0Q9W2V4 A0A3B0IYB5 A0A0P8YCY7 A0A0R3NQE0 A0A1I8NIA2 B4MIU5 A0A0B4KF89 A0A0M8ZTH3 A0A1Y1NDS6 A0A1Y1NHG0 B4P5X7 A0A1B6EB80 A0A1B0CSE2 B4KSN2 F4X0D9 B4LPA1 B3MDK1 A0A1W4X3C2 Q28WP0 A0A0L7R7G7 B4H762 A0A0P5L4T0 A0A0P6FVQ0 A0A0P5L219 A0A1W4UZI0 A0A0P5JMY7 O61366 B4HT72 B4QI16 A0A1D2NB16 W8AKW0
A0A182XD91 A0A182K8H4 A0A182PBN9 A0A182M4J2 A0A182NPH1 A0A182KSR9 A0A182Y1V7 A0A182QS62 A0A182GTH1 A0A084WLP8 E0VVS0 W5JQS1 A0A1Y1NDQ5 A0A182G5E8 A0A067RLF1 A0A182J907 A0A182HPZ9 A0A0J7L4B0 A0A1W4X3G2 A0A139WCU8 A0A336MNK9 A0A194Q1U0 A0A336MLJ2 A0A1W4XE57 E2BN74 Q7PV79 Q17M62 A0A1B0DHR1 A0A1I8PPP4 A0A151J7H1 A0A195D726 A0A1I8NIA6 K7IWI8 A0A0P9BZW6 A0A158NFL1 A0A0Q9XA06 A0A0R3NQU5 B0WEI5 A0A2A3EIW1 A0A0J9REF5 A0A0B4KFS9 V9I9S3 V9I9A5 A0A0Q5VLF4 A0A088AEG7 B4JWN0 A0A182FUE2 A0A1I8PPR4 A0A0L0BW10 A0A1A9X2G5 A0A182W989 A0A195AY50 A0A0Q9XAJ0 A0A0R3NQ29 A0A026WS64 A0A1A9XXV3 A0A0B4KEX5 A0A0R1DSS3 A0A0J9RE05 A0A1W4UZH5 A0A0Q5VM06 A0A1B6D7J1 A0A3L8DD70 A0A0Q9W2V4 A0A3B0IYB5 A0A0P8YCY7 A0A0R3NQE0 A0A1I8NIA2 B4MIU5 A0A0B4KF89 A0A0M8ZTH3 A0A1Y1NDS6 A0A1Y1NHG0 B4P5X7 A0A1B6EB80 A0A1B0CSE2 B4KSN2 F4X0D9 B4LPA1 B3MDK1 A0A1W4X3C2 Q28WP0 A0A0L7R7G7 B4H762 A0A0P5L4T0 A0A0P6FVQ0 A0A0P5L219 A0A1W4UZI0 A0A0P5JMY7 O61366 B4HT72 B4QI16 A0A1D2NB16 W8AKW0
EC Number
3.2.1.1
Pubmed
22118469
26354079
20966253
25244985
26483478
24438588
+ More
20566863 20920257 23761445 28004739 24845553 18362917 19820115 20798317 12364791 17510324 25315136 20075255 17994087 21347285 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26108605 18057021 23185243 24508170 17550304 30249741 21719571 27289101 24495485
20566863 20920257 23761445 28004739 24845553 18362917 19820115 20798317 12364791 17510324 25315136 20075255 17994087 21347285 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26108605 18057021 23185243 24508170 17550304 30249741 21719571 27289101 24495485
EMBL
ODYU01005621
SOQ46654.1
NWSH01002549
PCG68003.1
AGBW02014050
OWR42347.1
+ More
KQ459761 KPJ20142.1 AXCM01009750 AXCN02001829 JXUM01002040 KQ560132 KXJ84271.1 ATLV01024268 KE525351 KFB51142.1 DS235813 EEB17476.1 ADMH02000607 ETN65653.1 GEZM01005573 GEZM01005572 JAV96084.1 JXUM01007104 JXUM01007105 JXUM01007106 KQ560230 KXJ83722.1 KK852439 KDR23868.1 APCN01003285 LBMM01000800 KMQ97466.1 KQ971363 KYB25707.1 UFQT01001639 SSX31285.1 KQ459580 KPI99293.1 SSX31284.1 GL449385 EFN82836.1 AAAB01008986 EAA00303.5 CH477208 EAT47783.1 AJVK01015394 AJVK01015395 KQ979748 KYN19340.1 KQ976750 KYN08662.1 CH902619 KPU76843.1 ADTU01014616 CH933808 KRG05295.1 CM000071 KRT03244.1 DS231910 EDS45701.1 KZ288230 PBC31628.1 CM002911 KMY94311.1 AE013599 AGB93562.1 JR036551 AEY57221.1 JR036552 AEY57222.1 CH954179 KQS62432.1 CH916375 EDV98368.1 JRES01001254 KNC24211.1 KQ976716 KYM76869.1 KRG05292.1 KRG05293.1 KRG05296.1 KRT03240.1 KRT03242.1 KRT03246.1 KRT03247.1 KK107119 EZA58798.1 AGB93563.1 CM000158 KRK00185.1 KMY94312.1 KMY94313.1 KQS62434.1 KQS62435.1 KQS62436.1 GEDC01015656 JAS21642.1 QOIP01000009 RLU18256.1 CH940648 KRF79309.1 OUUW01000001 SPP72775.1 KPU76842.1 KRT03243.1 CH963719 EDW72034.2 AGB93560.1 KQ435855 KOX70627.1 GEZM01005575 GEZM01005574 JAV96082.1 GEZM01005571 JAV96085.1 EDW91893.2 GEDC01002117 JAS35181.1 AJWK01025918 AJWK01025919 EDW10531.1 GL888497 EGI60097.1 EDW60210.1 EDV37464.2 EAL26627.2 KQ414640 KOC66789.1 CH479216 EDW33595.1 GDIQ01199961 JAK51764.1 GDIQ01042128 JAN52609.1 GDIQ01201260 JAK50465.1 GDIQ01202593 JAK49132.1 AF022713 AAF57972.2 CH480816 EDW48173.1 CM000362 EDX07387.1 LJIJ01000111 ODN02448.1 GAMC01020102 GAMC01020100 GAMC01020098 GAMC01020096 JAB86457.1
KQ459761 KPJ20142.1 AXCM01009750 AXCN02001829 JXUM01002040 KQ560132 KXJ84271.1 ATLV01024268 KE525351 KFB51142.1 DS235813 EEB17476.1 ADMH02000607 ETN65653.1 GEZM01005573 GEZM01005572 JAV96084.1 JXUM01007104 JXUM01007105 JXUM01007106 KQ560230 KXJ83722.1 KK852439 KDR23868.1 APCN01003285 LBMM01000800 KMQ97466.1 KQ971363 KYB25707.1 UFQT01001639 SSX31285.1 KQ459580 KPI99293.1 SSX31284.1 GL449385 EFN82836.1 AAAB01008986 EAA00303.5 CH477208 EAT47783.1 AJVK01015394 AJVK01015395 KQ979748 KYN19340.1 KQ976750 KYN08662.1 CH902619 KPU76843.1 ADTU01014616 CH933808 KRG05295.1 CM000071 KRT03244.1 DS231910 EDS45701.1 KZ288230 PBC31628.1 CM002911 KMY94311.1 AE013599 AGB93562.1 JR036551 AEY57221.1 JR036552 AEY57222.1 CH954179 KQS62432.1 CH916375 EDV98368.1 JRES01001254 KNC24211.1 KQ976716 KYM76869.1 KRG05292.1 KRG05293.1 KRG05296.1 KRT03240.1 KRT03242.1 KRT03246.1 KRT03247.1 KK107119 EZA58798.1 AGB93563.1 CM000158 KRK00185.1 KMY94312.1 KMY94313.1 KQS62434.1 KQS62435.1 KQS62436.1 GEDC01015656 JAS21642.1 QOIP01000009 RLU18256.1 CH940648 KRF79309.1 OUUW01000001 SPP72775.1 KPU76842.1 KRT03243.1 CH963719 EDW72034.2 AGB93560.1 KQ435855 KOX70627.1 GEZM01005575 GEZM01005574 JAV96082.1 GEZM01005571 JAV96085.1 EDW91893.2 GEDC01002117 JAS35181.1 AJWK01025918 AJWK01025919 EDW10531.1 GL888497 EGI60097.1 EDW60210.1 EDV37464.2 EAL26627.2 KQ414640 KOC66789.1 CH479216 EDW33595.1 GDIQ01199961 JAK51764.1 GDIQ01042128 JAN52609.1 GDIQ01201260 JAK50465.1 GDIQ01202593 JAK49132.1 AF022713 AAF57972.2 CH480816 EDW48173.1 CM000362 EDX07387.1 LJIJ01000111 ODN02448.1 GAMC01020102 GAMC01020100 GAMC01020098 GAMC01020096 JAB86457.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000075900
UP000075903
UP000076407
+ More
UP000075881 UP000075885 UP000075883 UP000075884 UP000075882 UP000076408 UP000075886 UP000069940 UP000249989 UP000030765 UP000009046 UP000000673 UP000027135 UP000075880 UP000075840 UP000036403 UP000192223 UP000007266 UP000053268 UP000008237 UP000007062 UP000008820 UP000092462 UP000095300 UP000078492 UP000078542 UP000095301 UP000002358 UP000007801 UP000005205 UP000009192 UP000001819 UP000002320 UP000242457 UP000000803 UP000008711 UP000005203 UP000001070 UP000069272 UP000037069 UP000091820 UP000075920 UP000078540 UP000053097 UP000092443 UP000002282 UP000192221 UP000279307 UP000008792 UP000268350 UP000007798 UP000053105 UP000092461 UP000007755 UP000053825 UP000008744 UP000001292 UP000000304 UP000094527
UP000075881 UP000075885 UP000075883 UP000075884 UP000075882 UP000076408 UP000075886 UP000069940 UP000249989 UP000030765 UP000009046 UP000000673 UP000027135 UP000075880 UP000075840 UP000036403 UP000192223 UP000007266 UP000053268 UP000008237 UP000007062 UP000008820 UP000092462 UP000095300 UP000078492 UP000078542 UP000095301 UP000002358 UP000007801 UP000005205 UP000009192 UP000001819 UP000002320 UP000242457 UP000000803 UP000008711 UP000005203 UP000001070 UP000069272 UP000037069 UP000091820 UP000075920 UP000078540 UP000053097 UP000092443 UP000002282 UP000192221 UP000279307 UP000008792 UP000268350 UP000007798 UP000053105 UP000092461 UP000007755 UP000053825 UP000008744 UP000001292 UP000000304 UP000094527
PRIDE
Pfam
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2H1W2A6
A0A2A4J9C8
A0A212ELJ8
A0A0N1IQ65
A0A182R5H0
A0A182VLN3
+ More
A0A182XD91 A0A182K8H4 A0A182PBN9 A0A182M4J2 A0A182NPH1 A0A182KSR9 A0A182Y1V7 A0A182QS62 A0A182GTH1 A0A084WLP8 E0VVS0 W5JQS1 A0A1Y1NDQ5 A0A182G5E8 A0A067RLF1 A0A182J907 A0A182HPZ9 A0A0J7L4B0 A0A1W4X3G2 A0A139WCU8 A0A336MNK9 A0A194Q1U0 A0A336MLJ2 A0A1W4XE57 E2BN74 Q7PV79 Q17M62 A0A1B0DHR1 A0A1I8PPP4 A0A151J7H1 A0A195D726 A0A1I8NIA6 K7IWI8 A0A0P9BZW6 A0A158NFL1 A0A0Q9XA06 A0A0R3NQU5 B0WEI5 A0A2A3EIW1 A0A0J9REF5 A0A0B4KFS9 V9I9S3 V9I9A5 A0A0Q5VLF4 A0A088AEG7 B4JWN0 A0A182FUE2 A0A1I8PPR4 A0A0L0BW10 A0A1A9X2G5 A0A182W989 A0A195AY50 A0A0Q9XAJ0 A0A0R3NQ29 A0A026WS64 A0A1A9XXV3 A0A0B4KEX5 A0A0R1DSS3 A0A0J9RE05 A0A1W4UZH5 A0A0Q5VM06 A0A1B6D7J1 A0A3L8DD70 A0A0Q9W2V4 A0A3B0IYB5 A0A0P8YCY7 A0A0R3NQE0 A0A1I8NIA2 B4MIU5 A0A0B4KF89 A0A0M8ZTH3 A0A1Y1NDS6 A0A1Y1NHG0 B4P5X7 A0A1B6EB80 A0A1B0CSE2 B4KSN2 F4X0D9 B4LPA1 B3MDK1 A0A1W4X3C2 Q28WP0 A0A0L7R7G7 B4H762 A0A0P5L4T0 A0A0P6FVQ0 A0A0P5L219 A0A1W4UZI0 A0A0P5JMY7 O61366 B4HT72 B4QI16 A0A1D2NB16 W8AKW0
A0A182XD91 A0A182K8H4 A0A182PBN9 A0A182M4J2 A0A182NPH1 A0A182KSR9 A0A182Y1V7 A0A182QS62 A0A182GTH1 A0A084WLP8 E0VVS0 W5JQS1 A0A1Y1NDQ5 A0A182G5E8 A0A067RLF1 A0A182J907 A0A182HPZ9 A0A0J7L4B0 A0A1W4X3G2 A0A139WCU8 A0A336MNK9 A0A194Q1U0 A0A336MLJ2 A0A1W4XE57 E2BN74 Q7PV79 Q17M62 A0A1B0DHR1 A0A1I8PPP4 A0A151J7H1 A0A195D726 A0A1I8NIA6 K7IWI8 A0A0P9BZW6 A0A158NFL1 A0A0Q9XA06 A0A0R3NQU5 B0WEI5 A0A2A3EIW1 A0A0J9REF5 A0A0B4KFS9 V9I9S3 V9I9A5 A0A0Q5VLF4 A0A088AEG7 B4JWN0 A0A182FUE2 A0A1I8PPR4 A0A0L0BW10 A0A1A9X2G5 A0A182W989 A0A195AY50 A0A0Q9XAJ0 A0A0R3NQ29 A0A026WS64 A0A1A9XXV3 A0A0B4KEX5 A0A0R1DSS3 A0A0J9RE05 A0A1W4UZH5 A0A0Q5VM06 A0A1B6D7J1 A0A3L8DD70 A0A0Q9W2V4 A0A3B0IYB5 A0A0P8YCY7 A0A0R3NQE0 A0A1I8NIA2 B4MIU5 A0A0B4KF89 A0A0M8ZTH3 A0A1Y1NDS6 A0A1Y1NHG0 B4P5X7 A0A1B6EB80 A0A1B0CSE2 B4KSN2 F4X0D9 B4LPA1 B3MDK1 A0A1W4X3C2 Q28WP0 A0A0L7R7G7 B4H762 A0A0P5L4T0 A0A0P6FVQ0 A0A0P5L219 A0A1W4UZI0 A0A0P5JMY7 O61366 B4HT72 B4QI16 A0A1D2NB16 W8AKW0
PDB
6FFM
E-value=2.2877e-05,
Score=114
Ontologies
GO
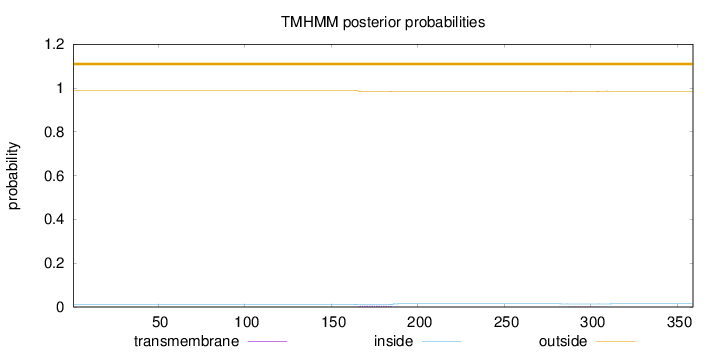
Topology
Length:
359
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13467
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01103
outside
1 - 359
Population Genetic Test Statistics
Pi
246.345391
Theta
184.01786
Tajima's D
0.510751
CLR
0.096029
CSRT
0.517674116294185
Interpretation
Uncertain
