Gene
KWMTBOMO04386 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005279
Annotation
PREDICTED:_putative_polypeptide_N-acetylgalactosaminyltransferase_9_isoform_X1_[Bombyx_mori]
Full name
Polypeptide N-acetylgalactosaminyltransferase
+ More
Putative polypeptide N-acetylgalactosaminyltransferase 9
Putative polypeptide N-acetylgalactosaminyltransferase 9
Alternative Name
Protein-UDP acetylgalactosaminyltransferase
Protein-UDP acetylgalactosaminyltransferase 9
UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 9
Protein-UDP acetylgalactosaminyltransferase 9
UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 9
Location in the cell
Cytoplasmic Reliability : 1.069 Mitochondrial Reliability : 1.08
Sequence
CDS
ATGAAATACTACAGCATGTTCTTCTTGAGGAGGCGAACCCTGATACTGAAAGCTGTTCTCATACTGACAGCTTTATGGTTTATGATAGCCCTGTTAACTACAAATGGCGGTTCACGGAACGCTATTGGCGCTCAGCCAGTTGAATACGAGGATGCTGAAGCGAAACCGTTGAAAATGGCCAAACCTACGTCGACAAGAAGGAAGAGTGAATCGTATGGCAACAATCTCTCGGACGGTCTAGGCGTGTTAGCTGCGCCCGGTTCGGACAATGCTCCCGGTGAACTTGGCAAACCAGTCGTACTGCCAAAGAACTTAACCGAAGAAACAAAACAGGCTGTCGCCGAGGGCTGGAAAAAGAACGCATTCAATCAATATGTGAGCGATCTTATATCGATCAGGAGGAAATTACCGGATCCTAGAGATGAGTGGTGCAAGCAACCTGGAAGGTATTTGGAAGATTTACCACAGACCTCGGTCGTAATATGCTTCCACAATGAAGCGTGGTCAGTGTTGTTGAGAACTGTGCATTCGGTTTTGGACCGATCACCAGCGCATCTGATCAAAGAAATCATACTTGTTGATGATTTTTCTGATATGCCACACTTGATGCAGCAACTGGACGATTATATGTCGTCACTCCCCAAGGTTAGAATAGTAAGAGCAACACGCCGTGAAGGCTTAATCAGGGCCAGATTGATCGGCGCCCGATACGTGACCGCCCCTATTCTAACCTATCTCGACAGTCATTGCGAATGTACTGAAGGTTGGCTGGAGCCCCTTCTTGATAGGATCGCGCGTAACAAAACGACCGTCGTCTGTCCTGTCATTGATGTAATTGATGACAATACTCTGGAGTATCACTATAGGGATTCTTCATCTGTCAACGTTGGTGGTTTCGACTGGAATTTACAGTTTAATTGGCATCCGGTACCATCGAAGGAACGCGCCAAGCATCAGAATACAGCAGAACCGGTTTGGTCTCCGACTATGGCCGGTGGCCTCTTTGCGATTGATAAGGAGTTCTTCGAAAAACTTGGAACCTACGATAACGGATTCGATATTTGGGGCGGTGAAAATTTGGAATTATCTTTTAAGACGTGGATGTGTGGAGGAACTCTTGAAATTGTGCCGTGCTCTCACGTTGGACATATTTTTAGAAAACGATCTCCATACAAGTGGCGAACAGGAGTTAATGTTTTAAAAAAGAATTCCGTGAGACTGGCAGAGGTTTGGCTCGATGATTACGCCAAATATTATTATCAACGTGTAGGTAACGATAAAGGGGACTACGGTGATGTCAGCAGCCGCAAAACACTGAGAAAAAATCTCGGCTGCAAATCTTTCGAATGGTACTTGAAAAACATCTATCCGGAATTGTTTATCCCTGGAGAATCGGTTGCTCATGGAGAGATACGCAATATTGGCTTCCAAAAGATTTGTCTGGACTCTCCAACCCGCAAGTCAGATCACCATAAGCCAGTCGGACTTTACCCGTGCCACCGACAGGGAGGAAATCAGTACTGGATGTATTCGAAAAACGGAGAGATACGTCGCGATGAAACTTGCTTGGACTATTCCGGGCACGATGTTGTATTGTACCCTTGTCACGGTGCTAAGGGAAATCAATTATGGTTGTACGATCCTAATACAAAGCTACTAAAACATGGCTCAAGCGAGAAATGCATGGCTATTTCTCGTAATAAGGACAAGATCACCATGGAAACGTGCAAAGAATCAGAAGACAGGCAGAAATGGACAATGGAGAATTTCAATGCTGAGCGCCTCAGTCCAGAACTTATAGCTGAGATCCAGTAG
Protein
MKYYSMFFLRRRTLILKAVLILTALWFMIALLTTNGGSRNAIGAQPVEYEDAEAKPLKMAKPTSTRRKSESYGNNLSDGLGVLAAPGSDNAPGELGKPVVLPKNLTEETKQAVAEGWKKNAFNQYVSDLISIRRKLPDPRDEWCKQPGRYLEDLPQTSVVICFHNEAWSVLLRTVHSVLDRSPAHLIKEIILVDDFSDMPHLMQQLDDYMSSLPKVRIVRATRREGLIRARLIGARYVTAPILTYLDSHCECTEGWLEPLLDRIARNKTTVVCPVIDVIDDNTLEYHYRDSSSVNVGGFDWNLQFNWHPVPSKERAKHQNTAEPVWSPTMAGGLFAIDKEFFEKLGTYDNGFDIWGGENLELSFKTWMCGGTLEIVPCSHVGHIFRKRSPYKWRTGVNVLKKNSVRLAEVWLDDYAKYYYQRVGNDKGDYGDVSSRKTLRKNLGCKSFEWYLKNIYPELFIPGESVAHGEIRNIGFQKICLDSPTRKSDHHKPVGLYPCHRQGGNQYWMYSKNGEIRRDETCLDYSGHDVVLYPCHGAKGNQLWLYDPNTKLLKHGSSEKCMAISRNKDKITMETCKESEDRQKWTMENFNAERLSPELIAEIQ
Summary
Description
May catalyze the initial reaction in O-linked oligosaccharide biosynthesis, the transfer of an N-acetyl-D-galactosamine residue to a serine or threonine residue on the protein receptor.
Catalytic Activity
L-seryl-[protein] + UDP-N-acetyl-alpha-D-galactosamine = 3-O-[N-acetyl-alpha-D-galactosaminyl]-L-seryl-[protein] + H(+) + UDP
L-threonyl-[protein] + UDP-N-acetyl-alpha-D-galactosamine = 3-O-[N-acetyl-alpha-D-galactosaminyl]-L-threonyl-[protein] + H(+) + UDP
L-threonyl-[protein] + UDP-N-acetyl-alpha-D-galactosamine = 3-O-[N-acetyl-alpha-D-galactosaminyl]-L-threonyl-[protein] + H(+) + UDP
Cofactor
Mn(2+)
Similarity
Belongs to the glycosyltransferase 2 family. GalNAc-T subfamily.
Keywords
3D-structure
Alternative splicing
Complete proteome
Disulfide bond
Glycoprotein
Glycosyltransferase
Golgi apparatus
Lectin
Manganese
Membrane
Metal-binding
Reference proteome
Signal-anchor
Transferase
Transmembrane
Transmembrane helix
Feature
chain Putative polypeptide N-acetylgalactosaminyltransferase 9
splice variant In isoform E.
splice variant In isoform E.
Uniprot
A0A3S2TPK5
A0A2A4K279
A0A2H1WZ65
A0A194Q2R8
A0A0N1PIE7
H9J6Y7
+ More
A0A212F578 A0A1Y1JSS5 A0A139WCV3 U5EVC4 A0A1Q3G2R0 A0A1Q3G2S0 A0A1Q3G2H7 A0A0K8VWG7 A0A034VQH2 Q17M64 A0A1B6CXC6 A0A182GTH0 A0A023EUU3 A0A2H8TZ85 A0A1I8MII7 A0A1L8E1X9 W8AHD0 A0A1L8E1J6 A0A0P4VZX2 A0A2A3EKC2 A0A158NFL3 A0A1B6D6D2 A0A034VRC2 A0A0C9QHK0 A0A2M4CQV6 A0A088AEG6 A0A0P4W672 D6WY29 A0A2M3Z2J8 E0VG07 K7IWI9 A0A2M4CPY5 A0A182FUE1 A0A2J7QSW7 A0A067RJ22 J9K3C1 A0A2M3YZD5 A0A2M4BG78 A0A3L8DCQ0 A0A1B6E8D1 A0A182K0L1 B4KSN8 A0A2M4A019 A0A182UF65 B4LP94 A0A2M4A0M0 A0A182VEZ2 A0A182KSS0 A0A182HQ00 A0A182XD90 A0A182R5G9 A0A146LKK0 B4QIE5 B4P5X0 B4MIT5 B4HT77 A0A1W4UXH9 W5JMP6 B3NP82 A0A182PBP0 B3MDJ4 A0A0A9WQI9 A0A1B6D832 Q8MRC9 A0A182QCF3 A0A182W988 A0A0Q9XJ23 A0A2R7WZE6 B4H754 A0A2I9LPG6 B5DZF4 A0A3B0IXZ9 A0A0P5KTU2 A0A0P6B1L1 A0A0A9WSG8 A0A0Q9W3D9 A0A151X532 A0A0P5ITQ0 A0A0P5DY35 A0A0Q9WYK2 A0A0R1DSH5 A0A0J9RE15 A0A1W4UKJ2 A0A0Q5VL32 A0A0K8WEC4 Q8MRC9-2 A0A1Z5KV28 A0A2L2Y6F0 B4JWN6 A0A1B0CSE3 A0A0N8P0D7 A0A3B0J1R1 A0A2R5LF21 A0A0R3NXN7
A0A212F578 A0A1Y1JSS5 A0A139WCV3 U5EVC4 A0A1Q3G2R0 A0A1Q3G2S0 A0A1Q3G2H7 A0A0K8VWG7 A0A034VQH2 Q17M64 A0A1B6CXC6 A0A182GTH0 A0A023EUU3 A0A2H8TZ85 A0A1I8MII7 A0A1L8E1X9 W8AHD0 A0A1L8E1J6 A0A0P4VZX2 A0A2A3EKC2 A0A158NFL3 A0A1B6D6D2 A0A034VRC2 A0A0C9QHK0 A0A2M4CQV6 A0A088AEG6 A0A0P4W672 D6WY29 A0A2M3Z2J8 E0VG07 K7IWI9 A0A2M4CPY5 A0A182FUE1 A0A2J7QSW7 A0A067RJ22 J9K3C1 A0A2M3YZD5 A0A2M4BG78 A0A3L8DCQ0 A0A1B6E8D1 A0A182K0L1 B4KSN8 A0A2M4A019 A0A182UF65 B4LP94 A0A2M4A0M0 A0A182VEZ2 A0A182KSS0 A0A182HQ00 A0A182XD90 A0A182R5G9 A0A146LKK0 B4QIE5 B4P5X0 B4MIT5 B4HT77 A0A1W4UXH9 W5JMP6 B3NP82 A0A182PBP0 B3MDJ4 A0A0A9WQI9 A0A1B6D832 Q8MRC9 A0A182QCF3 A0A182W988 A0A0Q9XJ23 A0A2R7WZE6 B4H754 A0A2I9LPG6 B5DZF4 A0A3B0IXZ9 A0A0P5KTU2 A0A0P6B1L1 A0A0A9WSG8 A0A0Q9W3D9 A0A151X532 A0A0P5ITQ0 A0A0P5DY35 A0A0Q9WYK2 A0A0R1DSH5 A0A0J9RE15 A0A1W4UKJ2 A0A0Q5VL32 A0A0K8WEC4 Q8MRC9-2 A0A1Z5KV28 A0A2L2Y6F0 B4JWN6 A0A1B0CSE3 A0A0N8P0D7 A0A3B0J1R1 A0A2R5LF21 A0A0R3NXN7
EC Number
2.4.1.-
2.4.1.41
2.4.1.41
Pubmed
26354079
19121390
22118469
28004739
18362917
19820115
+ More
25348373 17510324 26483478 24945155 25315136 24495485 21347285 20566863 20075255 24845553 30249741 17994087 18057021 20966253 26823975 22936249 17550304 20920257 23761445 25401762 10731132 12537572 12537569 16251381 29248469 15632085 23185243 28528879 26561354
25348373 17510324 26483478 24945155 25315136 24495485 21347285 20566863 20075255 24845553 30249741 17994087 18057021 20966253 26823975 22936249 17550304 20920257 23761445 25401762 10731132 12537572 12537569 16251381 29248469 15632085 23185243 28528879 26561354
EMBL
RSAL01000030
RVE51691.1
NWSH01000233
PCG78159.1
ODYU01012201
SOQ58380.1
+ More
KQ459580 KPI99294.1 KQ459761 KPJ20143.1 BABH01028458 AGBW02010253 OWR48859.1 GEZM01101764 GEZM01101763 GEZM01101762 GEZM01101761 GEZM01101759 GEZM01101758 JAV52322.1 KQ971362 KYB25760.1 GANO01003468 JAB56403.1 GFDL01001016 JAV34029.1 GFDL01001006 JAV34039.1 GFDL01001043 JAV34002.1 GDHF01009399 JAI42915.1 GAKP01014590 JAC44362.1 CH477208 EAT47781.1 GEDC01020658 GEDC01019191 GEDC01010305 JAS16640.1 JAS18107.1 JAS26993.1 JXUM01002040 KQ560132 KXJ84270.1 GAPW01000852 JAC12746.1 GFXV01007779 MBW19584.1 GFDF01001569 JAV12515.1 GAMC01021513 GAMC01021510 JAB85042.1 GFDF01001568 JAV12516.1 GDRN01083665 JAI61639.1 KZ288230 PBC31629.1 ADTU01014616 ADTU01014617 ADTU01014618 ADTU01014619 ADTU01014620 ADTU01014621 ADTU01014622 GEDC01016173 GEDC01004329 GEDC01001082 JAS21125.1 JAS32969.1 JAS36216.1 GAKP01014592 JAC44360.1 GBYB01003004 JAG72771.1 GGFL01003100 MBW67278.1 GDRN01083666 JAI61638.1 EFA07904.2 GGFM01001917 MBW22668.1 DS235129 EEB12313.1 GGFL01003073 MBW67251.1 NEVH01011211 PNF31680.1 KK852439 KDR23871.1 ABLF02041136 ABLF02041160 GGFM01000881 MBW21632.1 GGFJ01002886 MBW52027.1 QOIP01000009 RLU18255.1 GEDC01025537 GEDC01006763 GEDC01003108 JAS11761.1 JAS30535.1 JAS34190.1 CH933808 EDW10537.1 GGFK01000843 MBW34164.1 CH940648 EDW60203.2 KRF79303.1 GGFK01001033 MBW34354.1 APCN01003286 GDHC01010015 JAQ08614.1 CM000362 CM002911 EDX07392.1 KMY94322.1 CM000158 EDW91886.2 CH963719 EDW72024.1 CH480816 EDW48178.1 ADMH02000607 ETN65652.1 CH954179 EDV55721.2 CH902619 EDV37457.2 GBHO01032877 GBHO01032876 JAG10727.1 JAG10728.1 GEDC01015957 GEDC01015451 GEDC01000234 JAS21341.1 JAS21847.1 JAS37064.1 AE013599 AY121661 BT029588 BT029630 AXCN02001829 KRG05300.1 KK856105 PTY24892.1 CH479216 EDW33587.1 GFWZ01000290 MBW20280.1 CM000071 EDY69575.1 KRT03239.1 OUUW01000001 SPP72784.1 GDIQ01205154 JAK46571.1 GDIP01021717 JAM81998.1 GBHO01032875 GBHO01032874 GBRD01015740 GBRD01015738 JAG10729.1 JAG10730.1 JAG50086.1 KRF79302.1 KQ982536 KYQ55434.1 GDIQ01209208 JAK42517.1 GDIP01155548 JAJ67854.1 KRF97468.1 KRK00182.1 KMY94323.1 KQS62429.1 GDHF01002925 JAI49389.1 GFJQ02008226 JAV98743.1 IAAA01004297 IAAA01004298 IAAA01004299 IAAA01004300 IAAA01004301 LAA02725.1 CH916375 EDV98374.1 AJWK01025919 KPU76836.1 SPP72783.1 GGLE01003995 MBY08121.1 KRT03238.1
KQ459580 KPI99294.1 KQ459761 KPJ20143.1 BABH01028458 AGBW02010253 OWR48859.1 GEZM01101764 GEZM01101763 GEZM01101762 GEZM01101761 GEZM01101759 GEZM01101758 JAV52322.1 KQ971362 KYB25760.1 GANO01003468 JAB56403.1 GFDL01001016 JAV34029.1 GFDL01001006 JAV34039.1 GFDL01001043 JAV34002.1 GDHF01009399 JAI42915.1 GAKP01014590 JAC44362.1 CH477208 EAT47781.1 GEDC01020658 GEDC01019191 GEDC01010305 JAS16640.1 JAS18107.1 JAS26993.1 JXUM01002040 KQ560132 KXJ84270.1 GAPW01000852 JAC12746.1 GFXV01007779 MBW19584.1 GFDF01001569 JAV12515.1 GAMC01021513 GAMC01021510 JAB85042.1 GFDF01001568 JAV12516.1 GDRN01083665 JAI61639.1 KZ288230 PBC31629.1 ADTU01014616 ADTU01014617 ADTU01014618 ADTU01014619 ADTU01014620 ADTU01014621 ADTU01014622 GEDC01016173 GEDC01004329 GEDC01001082 JAS21125.1 JAS32969.1 JAS36216.1 GAKP01014592 JAC44360.1 GBYB01003004 JAG72771.1 GGFL01003100 MBW67278.1 GDRN01083666 JAI61638.1 EFA07904.2 GGFM01001917 MBW22668.1 DS235129 EEB12313.1 GGFL01003073 MBW67251.1 NEVH01011211 PNF31680.1 KK852439 KDR23871.1 ABLF02041136 ABLF02041160 GGFM01000881 MBW21632.1 GGFJ01002886 MBW52027.1 QOIP01000009 RLU18255.1 GEDC01025537 GEDC01006763 GEDC01003108 JAS11761.1 JAS30535.1 JAS34190.1 CH933808 EDW10537.1 GGFK01000843 MBW34164.1 CH940648 EDW60203.2 KRF79303.1 GGFK01001033 MBW34354.1 APCN01003286 GDHC01010015 JAQ08614.1 CM000362 CM002911 EDX07392.1 KMY94322.1 CM000158 EDW91886.2 CH963719 EDW72024.1 CH480816 EDW48178.1 ADMH02000607 ETN65652.1 CH954179 EDV55721.2 CH902619 EDV37457.2 GBHO01032877 GBHO01032876 JAG10727.1 JAG10728.1 GEDC01015957 GEDC01015451 GEDC01000234 JAS21341.1 JAS21847.1 JAS37064.1 AE013599 AY121661 BT029588 BT029630 AXCN02001829 KRG05300.1 KK856105 PTY24892.1 CH479216 EDW33587.1 GFWZ01000290 MBW20280.1 CM000071 EDY69575.1 KRT03239.1 OUUW01000001 SPP72784.1 GDIQ01205154 JAK46571.1 GDIP01021717 JAM81998.1 GBHO01032875 GBHO01032874 GBRD01015740 GBRD01015738 JAG10729.1 JAG10730.1 JAG50086.1 KRF79302.1 KQ982536 KYQ55434.1 GDIQ01209208 JAK42517.1 GDIP01155548 JAJ67854.1 KRF97468.1 KRK00182.1 KMY94323.1 KQS62429.1 GDHF01002925 JAI49389.1 GFJQ02008226 JAV98743.1 IAAA01004297 IAAA01004298 IAAA01004299 IAAA01004300 IAAA01004301 LAA02725.1 CH916375 EDV98374.1 AJWK01025919 KPU76836.1 SPP72783.1 GGLE01003995 MBY08121.1 KRT03238.1
Proteomes
UP000283053
UP000218220
UP000053268
UP000053240
UP000005204
UP000007151
+ More
UP000007266 UP000008820 UP000069940 UP000249989 UP000095301 UP000242457 UP000005205 UP000005203 UP000009046 UP000002358 UP000069272 UP000235965 UP000027135 UP000007819 UP000279307 UP000075881 UP000009192 UP000075902 UP000008792 UP000075903 UP000075882 UP000075840 UP000076407 UP000075900 UP000000304 UP000002282 UP000007798 UP000001292 UP000192221 UP000000673 UP000008711 UP000075885 UP000007801 UP000000803 UP000075886 UP000075920 UP000008744 UP000001819 UP000268350 UP000075809 UP000001070 UP000092461
UP000007266 UP000008820 UP000069940 UP000249989 UP000095301 UP000242457 UP000005205 UP000005203 UP000009046 UP000002358 UP000069272 UP000235965 UP000027135 UP000007819 UP000279307 UP000075881 UP000009192 UP000075902 UP000008792 UP000075903 UP000075882 UP000075840 UP000076407 UP000075900 UP000000304 UP000002282 UP000007798 UP000001292 UP000192221 UP000000673 UP000008711 UP000075885 UP000007801 UP000000803 UP000075886 UP000075920 UP000008744 UP000001819 UP000268350 UP000075809 UP000001070 UP000092461
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A3S2TPK5
A0A2A4K279
A0A2H1WZ65
A0A194Q2R8
A0A0N1PIE7
H9J6Y7
+ More
A0A212F578 A0A1Y1JSS5 A0A139WCV3 U5EVC4 A0A1Q3G2R0 A0A1Q3G2S0 A0A1Q3G2H7 A0A0K8VWG7 A0A034VQH2 Q17M64 A0A1B6CXC6 A0A182GTH0 A0A023EUU3 A0A2H8TZ85 A0A1I8MII7 A0A1L8E1X9 W8AHD0 A0A1L8E1J6 A0A0P4VZX2 A0A2A3EKC2 A0A158NFL3 A0A1B6D6D2 A0A034VRC2 A0A0C9QHK0 A0A2M4CQV6 A0A088AEG6 A0A0P4W672 D6WY29 A0A2M3Z2J8 E0VG07 K7IWI9 A0A2M4CPY5 A0A182FUE1 A0A2J7QSW7 A0A067RJ22 J9K3C1 A0A2M3YZD5 A0A2M4BG78 A0A3L8DCQ0 A0A1B6E8D1 A0A182K0L1 B4KSN8 A0A2M4A019 A0A182UF65 B4LP94 A0A2M4A0M0 A0A182VEZ2 A0A182KSS0 A0A182HQ00 A0A182XD90 A0A182R5G9 A0A146LKK0 B4QIE5 B4P5X0 B4MIT5 B4HT77 A0A1W4UXH9 W5JMP6 B3NP82 A0A182PBP0 B3MDJ4 A0A0A9WQI9 A0A1B6D832 Q8MRC9 A0A182QCF3 A0A182W988 A0A0Q9XJ23 A0A2R7WZE6 B4H754 A0A2I9LPG6 B5DZF4 A0A3B0IXZ9 A0A0P5KTU2 A0A0P6B1L1 A0A0A9WSG8 A0A0Q9W3D9 A0A151X532 A0A0P5ITQ0 A0A0P5DY35 A0A0Q9WYK2 A0A0R1DSH5 A0A0J9RE15 A0A1W4UKJ2 A0A0Q5VL32 A0A0K8WEC4 Q8MRC9-2 A0A1Z5KV28 A0A2L2Y6F0 B4JWN6 A0A1B0CSE3 A0A0N8P0D7 A0A3B0J1R1 A0A2R5LF21 A0A0R3NXN7
A0A212F578 A0A1Y1JSS5 A0A139WCV3 U5EVC4 A0A1Q3G2R0 A0A1Q3G2S0 A0A1Q3G2H7 A0A0K8VWG7 A0A034VQH2 Q17M64 A0A1B6CXC6 A0A182GTH0 A0A023EUU3 A0A2H8TZ85 A0A1I8MII7 A0A1L8E1X9 W8AHD0 A0A1L8E1J6 A0A0P4VZX2 A0A2A3EKC2 A0A158NFL3 A0A1B6D6D2 A0A034VRC2 A0A0C9QHK0 A0A2M4CQV6 A0A088AEG6 A0A0P4W672 D6WY29 A0A2M3Z2J8 E0VG07 K7IWI9 A0A2M4CPY5 A0A182FUE1 A0A2J7QSW7 A0A067RJ22 J9K3C1 A0A2M3YZD5 A0A2M4BG78 A0A3L8DCQ0 A0A1B6E8D1 A0A182K0L1 B4KSN8 A0A2M4A019 A0A182UF65 B4LP94 A0A2M4A0M0 A0A182VEZ2 A0A182KSS0 A0A182HQ00 A0A182XD90 A0A182R5G9 A0A146LKK0 B4QIE5 B4P5X0 B4MIT5 B4HT77 A0A1W4UXH9 W5JMP6 B3NP82 A0A182PBP0 B3MDJ4 A0A0A9WQI9 A0A1B6D832 Q8MRC9 A0A182QCF3 A0A182W988 A0A0Q9XJ23 A0A2R7WZE6 B4H754 A0A2I9LPG6 B5DZF4 A0A3B0IXZ9 A0A0P5KTU2 A0A0P6B1L1 A0A0A9WSG8 A0A0Q9W3D9 A0A151X532 A0A0P5ITQ0 A0A0P5DY35 A0A0Q9WYK2 A0A0R1DSH5 A0A0J9RE15 A0A1W4UKJ2 A0A0Q5VL32 A0A0K8WEC4 Q8MRC9-2 A0A1Z5KV28 A0A2L2Y6F0 B4JWN6 A0A1B0CSE3 A0A0N8P0D7 A0A3B0J1R1 A0A2R5LF21 A0A0R3NXN7
PDB
6E4Q
E-value=0,
Score=2145
Ontologies
PATHWAY
GO
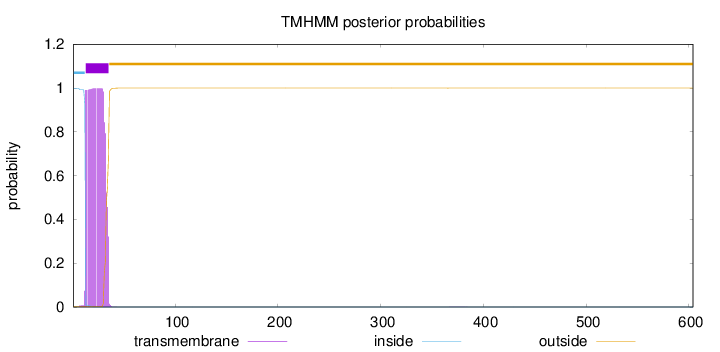
Topology
Subcellular location
Golgi apparatus membrane
Length:
604
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.00064
Exp number, first 60 AAs:
20.99793
Total prob of N-in:
0.99885
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 604
Population Genetic Test Statistics
Pi
229.321017
Theta
192.551715
Tajima's D
0.535067
CLR
0.006417
CSRT
0.51872406379681
Interpretation
Uncertain
