Gene
KWMTBOMO04385 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005278
Annotation
cuticular_protein_RR-1_motif_3_precursor_[Bombyx_mori]
Full name
Larval cuticle protein LCP-17
Location in the cell
Extracellular Reliability : 2.482
Sequence
CDS
ATGAGAACTTACATCTTCCTCGCTTTGGTCGCGGTAGCCGTCGCTGACGTTTCTCACGTCGTGCGCACTGGCGAAGCTGATGCCCAAATTGTGTCGCAAGATGCTGACGTTTTCCCGGACAAATACCAGTACCAATATCAGACCAGCAACGGAATCAGTGGCCAGGAACAAGGAGCGCTCGTAAACGAGGGCCGTGAGGATGCATCCATCGCCGTCCAAGGTTCAAGCGGCTACACTGCCCCTGATGGTACTCCCATTCAGATCACTTACATTGCTGACGCCAACGGATACCAGCCTTCGGGTGCCCATCTGCCCACTACCCCAGCCCCTGTCCCAATCCCAGATTACATCGCCCGGGCCATCGAGTACATCAGAACTCATCCACCCAAGCCAGAGGTCGGCCAACGTATTTAA
Protein
MRTYIFLALVAVAVADVSHVVRTGEADAQIVSQDADVFPDKYQYQYQTSNGISGQEQGALVNEGREDASIAVQGSSGYTAPDGTPIQITYIADANGYQPSGAHLPTTPAPVPIPDYIARAIEYIRTHPPKPEVGQRI
Summary
Keywords
Complete proteome
Cuticle
Reference proteome
Signal
Feature
chain Larval cuticle protein LCP-17
Uniprot
C0H6J4
H9J6Y6
A0A3S2M5W3
A0A0N1PIW1
I4DLT5
B2DBG7
+ More
A0A194Q7B5 A0A2H1W5N1 A0A212FID0 A0A2A4K2K2 I4DMG1 A0A0N1PJL0 I4DIL1 A0A194Q228 H9J6Y5 C0H6J3 W0GHP6 O02387 A0A0T6BD43 A0A3S2NH44 A0A2H1VH13 A0A1E1W218 A0A2A4IY21 S4S7G1 B2DBP1 C0H6N4 A0A2S1ZS80 A0A2W1CJG5 A0A1L8E3K8 R4G2S2 A0A182PN99 R4FKX5 T1I377 A0A0N1I9F1 A0A0K8SA91 I4DLT6 G8FVP6 A0A2A4K1N4 C0H6N5 A0A194Q164 A0A194R0P7 A0A067R439 A0A2H1X3S9 A0A194PX52 A0A2J7R753 A0A194R2A2 C0H6M4 A0A2H1VDB8 A0A182QNQ5 A0A0M4EMT1 A0A224XS48 A0A182S6R8 A0A1B0CM08 A0A1B0DQJ1 A0A182Y0U3 A0A182T445 A0A182HM56 C0H6J7 A0A0A1XLW3 A0A1Q3FZL7 A0A182LJP8 A0A182XCT0 Q7PNL5 V5IB19 A0A182RQ98 B0WPT5 A0A182IL85 Q7Q1A8 A0A182KTN1 A0A182U3Y3 A0A182VEM2 A0A182J1I2 W5JR27 A0A182U0L8 A0A2A4J2I4 A0A2S1ZS73 A0A1J1II08 B3M5M1 A0A1W4XNY4 A0A182MFL3 B3NCP1 A0A182RHG2 A0A034WSY4 A0A1W4XN34 A0A0L7LEM5 A0A084W2U4 K7IZX0 A0A182GEJ6 A0A182MJQ4 A0A182FFD4 A0A212F0M0 A0A182NMA0 K7IZX1 B4PEI4 A0A182YDM4 A0A182Q3H0
A0A194Q7B5 A0A2H1W5N1 A0A212FID0 A0A2A4K2K2 I4DMG1 A0A0N1PJL0 I4DIL1 A0A194Q228 H9J6Y5 C0H6J3 W0GHP6 O02387 A0A0T6BD43 A0A3S2NH44 A0A2H1VH13 A0A1E1W218 A0A2A4IY21 S4S7G1 B2DBP1 C0H6N4 A0A2S1ZS80 A0A2W1CJG5 A0A1L8E3K8 R4G2S2 A0A182PN99 R4FKX5 T1I377 A0A0N1I9F1 A0A0K8SA91 I4DLT6 G8FVP6 A0A2A4K1N4 C0H6N5 A0A194Q164 A0A194R0P7 A0A067R439 A0A2H1X3S9 A0A194PX52 A0A2J7R753 A0A194R2A2 C0H6M4 A0A2H1VDB8 A0A182QNQ5 A0A0M4EMT1 A0A224XS48 A0A182S6R8 A0A1B0CM08 A0A1B0DQJ1 A0A182Y0U3 A0A182T445 A0A182HM56 C0H6J7 A0A0A1XLW3 A0A1Q3FZL7 A0A182LJP8 A0A182XCT0 Q7PNL5 V5IB19 A0A182RQ98 B0WPT5 A0A182IL85 Q7Q1A8 A0A182KTN1 A0A182U3Y3 A0A182VEM2 A0A182J1I2 W5JR27 A0A182U0L8 A0A2A4J2I4 A0A2S1ZS73 A0A1J1II08 B3M5M1 A0A1W4XNY4 A0A182MFL3 B3NCP1 A0A182RHG2 A0A034WSY4 A0A1W4XN34 A0A0L7LEM5 A0A084W2U4 K7IZX0 A0A182GEJ6 A0A182MJQ4 A0A182FFD4 A0A212F0M0 A0A182NMA0 K7IZX1 B4PEI4 A0A182YDM4 A0A182Q3H0
Pubmed
EMBL
BR000504
FAA00505.1
BABH01028447
RSAL01000030
RVE51692.1
KQ460571
+ More
KPJ13960.1 AK402253 BAM18875.1 AB264646 BAG30726.1 KQ459580 KPI99295.1 ODYU01006181 SOQ47814.1 AGBW02008397 OWR53494.1 NWSH01000233 PCG78158.1 AK402479 BAM19101.1 KQ460400 KPJ14975.1 AK401129 BAM17751.1 KQ459564 KPI99622.1 BR000503 FAA00504.1 KF672849 AHF55751.1 AB004766 LJIG01001715 KRT85225.1 RSAL01000041 RVE50919.1 ODYU01002513 SOQ40106.1 GDQN01010014 JAT81040.1 NWSH01005682 PCG64060.1 JF415082 AFC88815.1 AB264726 BAG30800.1 BABH01013074 BR000543 FAA00545.1 MF942791 AWK28314.1 NHMN01024364 PZC87412.1 GFDF01000761 JAV13323.1 GAHY01002351 JAA75159.1 GAHY01002329 JAA75181.1 ACPB03023185 KPJ13959.1 GBRD01016156 JAG49670.1 AK402254 BAM18876.1 JN082716 AER27814.1 PCG78157.1 BABH01013076 BR000544 FAA00546.1 KPI99296.1 KQ460883 KPJ11368.1 KK852994 KDR12717.1 ODYU01013214 SOQ59908.1 KQ459586 KPI97902.1 NEVH01006738 PNF36675.1 KPJ11380.1 BABH01013051 BABH01013052 BR000533 FAA00535.1 ODYU01001934 SOQ38825.1 AXCN02002061 CP012525 ALC45072.1 GFTR01002482 JAW13944.1 AJWK01017996 AJVK01008596 APCN01004355 BABH01028739 BABH01028740 BR000507 FAA00508.1 GBXI01002286 JAD12006.1 GFDL01002058 JAV32987.1 AAAB01008960 EAA11968.4 GALX01000205 JAB68261.1 DS232029 EDS32500.1 AAAB01008980 EAA14443.4 ADMH02000355 ETN66852.1 NWSH01003986 PCG65593.1 MF942789 AWK28312.1 CVRI01000054 CRK99895.1 CH902618 EDV40655.1 AXCM01002738 CH954178 EDV51338.1 GAKP01001667 JAC57285.1 JTDY01001457 KOB73824.1 ATLV01019697 KE525278 KFB44538.1 JXUM01058156 JXUM01058157 KQ561989 KXJ76980.1 AXCM01001822 AGBW02011079 OWR47244.1 CM000159 EDW94050.1 AXCN02000938
KPJ13960.1 AK402253 BAM18875.1 AB264646 BAG30726.1 KQ459580 KPI99295.1 ODYU01006181 SOQ47814.1 AGBW02008397 OWR53494.1 NWSH01000233 PCG78158.1 AK402479 BAM19101.1 KQ460400 KPJ14975.1 AK401129 BAM17751.1 KQ459564 KPI99622.1 BR000503 FAA00504.1 KF672849 AHF55751.1 AB004766 LJIG01001715 KRT85225.1 RSAL01000041 RVE50919.1 ODYU01002513 SOQ40106.1 GDQN01010014 JAT81040.1 NWSH01005682 PCG64060.1 JF415082 AFC88815.1 AB264726 BAG30800.1 BABH01013074 BR000543 FAA00545.1 MF942791 AWK28314.1 NHMN01024364 PZC87412.1 GFDF01000761 JAV13323.1 GAHY01002351 JAA75159.1 GAHY01002329 JAA75181.1 ACPB03023185 KPJ13959.1 GBRD01016156 JAG49670.1 AK402254 BAM18876.1 JN082716 AER27814.1 PCG78157.1 BABH01013076 BR000544 FAA00546.1 KPI99296.1 KQ460883 KPJ11368.1 KK852994 KDR12717.1 ODYU01013214 SOQ59908.1 KQ459586 KPI97902.1 NEVH01006738 PNF36675.1 KPJ11380.1 BABH01013051 BABH01013052 BR000533 FAA00535.1 ODYU01001934 SOQ38825.1 AXCN02002061 CP012525 ALC45072.1 GFTR01002482 JAW13944.1 AJWK01017996 AJVK01008596 APCN01004355 BABH01028739 BABH01028740 BR000507 FAA00508.1 GBXI01002286 JAD12006.1 GFDL01002058 JAV32987.1 AAAB01008960 EAA11968.4 GALX01000205 JAB68261.1 DS232029 EDS32500.1 AAAB01008980 EAA14443.4 ADMH02000355 ETN66852.1 NWSH01003986 PCG65593.1 MF942789 AWK28312.1 CVRI01000054 CRK99895.1 CH902618 EDV40655.1 AXCM01002738 CH954178 EDV51338.1 GAKP01001667 JAC57285.1 JTDY01001457 KOB73824.1 ATLV01019697 KE525278 KFB44538.1 JXUM01058156 JXUM01058157 KQ561989 KXJ76980.1 AXCM01001822 AGBW02011079 OWR47244.1 CM000159 EDW94050.1 AXCN02000938
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000007151
UP000218220
+ More
UP000075885 UP000015103 UP000027135 UP000235965 UP000075886 UP000092553 UP000075901 UP000092461 UP000092462 UP000076408 UP000075840 UP000075882 UP000076407 UP000007062 UP000075900 UP000002320 UP000075880 UP000075902 UP000075903 UP000000673 UP000183832 UP000007801 UP000192223 UP000075883 UP000008711 UP000037510 UP000030765 UP000002358 UP000069940 UP000249989 UP000069272 UP000075884 UP000002282
UP000075885 UP000015103 UP000027135 UP000235965 UP000075886 UP000092553 UP000075901 UP000092461 UP000092462 UP000076408 UP000075840 UP000075882 UP000076407 UP000007062 UP000075900 UP000002320 UP000075880 UP000075902 UP000075903 UP000000673 UP000183832 UP000007801 UP000192223 UP000075883 UP000008711 UP000037510 UP000030765 UP000002358 UP000069940 UP000249989 UP000069272 UP000075884 UP000002282
Pfam
PF00379 Chitin_bind_4
ProteinModelPortal
C0H6J4
H9J6Y6
A0A3S2M5W3
A0A0N1PIW1
I4DLT5
B2DBG7
+ More
A0A194Q7B5 A0A2H1W5N1 A0A212FID0 A0A2A4K2K2 I4DMG1 A0A0N1PJL0 I4DIL1 A0A194Q228 H9J6Y5 C0H6J3 W0GHP6 O02387 A0A0T6BD43 A0A3S2NH44 A0A2H1VH13 A0A1E1W218 A0A2A4IY21 S4S7G1 B2DBP1 C0H6N4 A0A2S1ZS80 A0A2W1CJG5 A0A1L8E3K8 R4G2S2 A0A182PN99 R4FKX5 T1I377 A0A0N1I9F1 A0A0K8SA91 I4DLT6 G8FVP6 A0A2A4K1N4 C0H6N5 A0A194Q164 A0A194R0P7 A0A067R439 A0A2H1X3S9 A0A194PX52 A0A2J7R753 A0A194R2A2 C0H6M4 A0A2H1VDB8 A0A182QNQ5 A0A0M4EMT1 A0A224XS48 A0A182S6R8 A0A1B0CM08 A0A1B0DQJ1 A0A182Y0U3 A0A182T445 A0A182HM56 C0H6J7 A0A0A1XLW3 A0A1Q3FZL7 A0A182LJP8 A0A182XCT0 Q7PNL5 V5IB19 A0A182RQ98 B0WPT5 A0A182IL85 Q7Q1A8 A0A182KTN1 A0A182U3Y3 A0A182VEM2 A0A182J1I2 W5JR27 A0A182U0L8 A0A2A4J2I4 A0A2S1ZS73 A0A1J1II08 B3M5M1 A0A1W4XNY4 A0A182MFL3 B3NCP1 A0A182RHG2 A0A034WSY4 A0A1W4XN34 A0A0L7LEM5 A0A084W2U4 K7IZX0 A0A182GEJ6 A0A182MJQ4 A0A182FFD4 A0A212F0M0 A0A182NMA0 K7IZX1 B4PEI4 A0A182YDM4 A0A182Q3H0
A0A194Q7B5 A0A2H1W5N1 A0A212FID0 A0A2A4K2K2 I4DMG1 A0A0N1PJL0 I4DIL1 A0A194Q228 H9J6Y5 C0H6J3 W0GHP6 O02387 A0A0T6BD43 A0A3S2NH44 A0A2H1VH13 A0A1E1W218 A0A2A4IY21 S4S7G1 B2DBP1 C0H6N4 A0A2S1ZS80 A0A2W1CJG5 A0A1L8E3K8 R4G2S2 A0A182PN99 R4FKX5 T1I377 A0A0N1I9F1 A0A0K8SA91 I4DLT6 G8FVP6 A0A2A4K1N4 C0H6N5 A0A194Q164 A0A194R0P7 A0A067R439 A0A2H1X3S9 A0A194PX52 A0A2J7R753 A0A194R2A2 C0H6M4 A0A2H1VDB8 A0A182QNQ5 A0A0M4EMT1 A0A224XS48 A0A182S6R8 A0A1B0CM08 A0A1B0DQJ1 A0A182Y0U3 A0A182T445 A0A182HM56 C0H6J7 A0A0A1XLW3 A0A1Q3FZL7 A0A182LJP8 A0A182XCT0 Q7PNL5 V5IB19 A0A182RQ98 B0WPT5 A0A182IL85 Q7Q1A8 A0A182KTN1 A0A182U3Y3 A0A182VEM2 A0A182J1I2 W5JR27 A0A182U0L8 A0A2A4J2I4 A0A2S1ZS73 A0A1J1II08 B3M5M1 A0A1W4XNY4 A0A182MFL3 B3NCP1 A0A182RHG2 A0A034WSY4 A0A1W4XN34 A0A0L7LEM5 A0A084W2U4 K7IZX0 A0A182GEJ6 A0A182MJQ4 A0A182FFD4 A0A212F0M0 A0A182NMA0 K7IZX1 B4PEI4 A0A182YDM4 A0A182Q3H0
Ontologies
GO
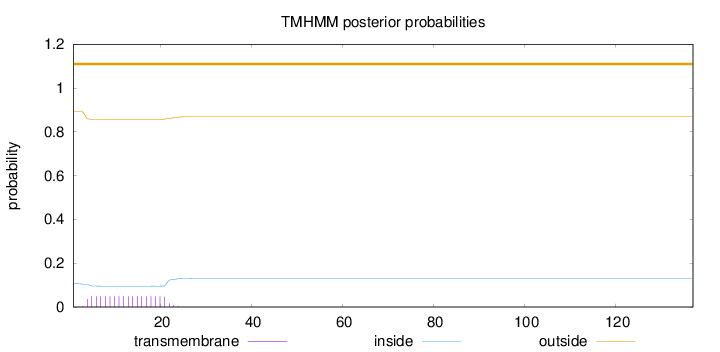
Topology
SignalP
Position: 1 - 15,
Likelihood: 0.994350
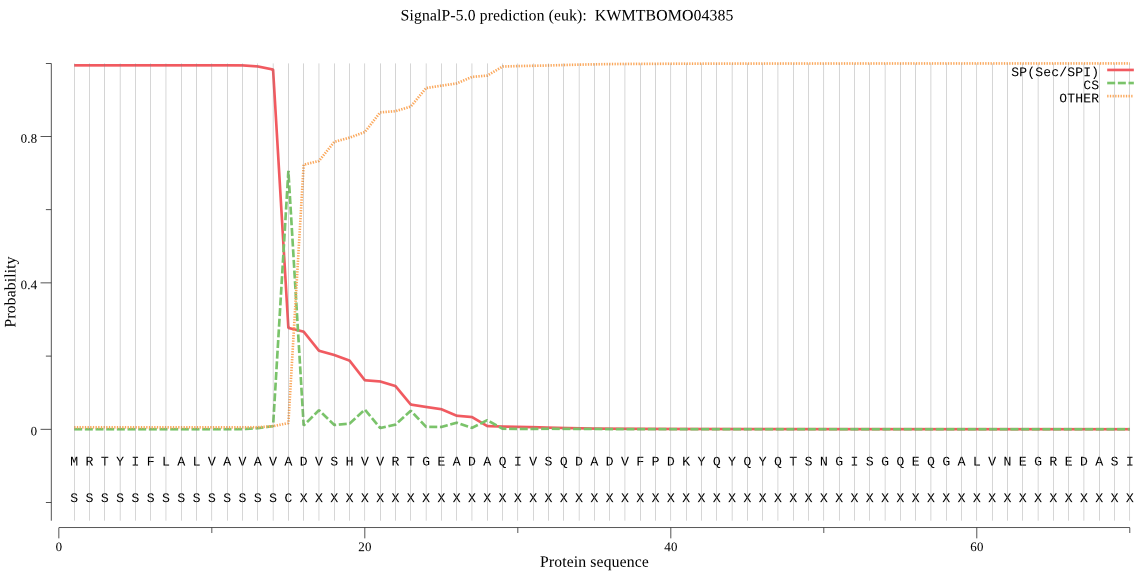
Length:
137
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.899949999999999
Exp number, first 60 AAs:
0.89966
Total prob of N-in:
0.10814
outside
1 - 137
Population Genetic Test Statistics
Pi
215.09001
Theta
145.459149
Tajima's D
1.448268
CLR
0.237963
CSRT
0.769511524423779
Interpretation
Uncertain
