Gene
KWMTBOMO04380
Pre Gene Modal
BGIBMGA005275
Annotation
PREDICTED:_PAB-dependent_poly(A)-specific_ribonuclease_subunit_PAN2_isoform_X2_[Bombyx_mori]
Full name
PAN2-PAN3 deadenylation complex catalytic subunit PAN2
Alternative Name
PAB1P-dependent poly(A)-specific ribonuclease
Poly(A)-nuclease deadenylation complex subunit 2
Poly(A)-nuclease deadenylation complex subunit 2
Location in the cell
Cytoplasmic Reliability : 1.3 Nuclear Reliability : 1.971
Sequence
CDS
ATGGAATATCATTACTCCGAAATATGTATGCCATCCGATCATCTAACAGATTACACACCCCCTTTCATCATTCCCACGCATGAAGGGGAATATGAAGCACGTAACAGTGTACTCGTGGATGGAGGAGAGAGATTTGGAGTTTCGGCAGTTGTGTTCGATAAACATGAAGAGTTGATATGGATGGGGAATCAGGGTGGTCATGTAACATCCTACTACGGTCCAAATATGCAAAAGTACACATCATTTCAAGTACACGATACTGATGAAGTTAGAGATATTATCACTGTTGAAAAATGTATTTATGTTTTGACCAAAACAACTCTACGCCATCAGTTACGTAGAGGTATACCTAAGCACACGCACAAATCACCAAACATGACAGACATGCAATGCCTGTATCAAGTGAACTCTACAAAATTATTGATGGGCGGACACCAAGACAAACTAATCGATTTGGACCTTAATAAAATGGTTGAAACTATCATTCCAATACAAGAGGAAGGCTGTGCGGTATTAAGAAGCGGTGGTAGCAGTCACGTCGCTTGTGGTAGTGCTAACGGTATAGTGGCATTGCGGGATATGCGTGCTCCTACAAAAGCTGAGCATACATTCAGAGCACACACGGCTTGTCTATCTGATCTGGATATGCAAGGAGATCTGTTGCTCACATGTGGATTTACTCAGGCCGTTGGAGGGGTAGTCGCAGAACCCTACATAGTGGTCTGGGACGTCCGATGTCTACGCGGCAGTAACCCTGGACAAGGAGGTTGGACCATTCATACCACATCACCGCCACTGCTACTTCACTTCCTGCCGGCGTTCTCTGGTAGGGCAGTAGCTGCTTCGGGCGACGGACACGTCAGTTTGTTACACGTGAACGCACCTGAAGAAAGGCAGTCAATTTTTAAGGTAGAGACGCACAGTTCACAGTGCAATGTGATGGACGTTGCGTCGACGAGTCAAGCCCTCGCTTTCGGCGACCAATCAGGACATTTAAATCTTTTCTCACCGACGCAGAATCACGAACCTGTATACAATAATTTTTCAAGGTCAACAGAGTTCGCGGATCCAGTGGAAGGTCTACCTTACGTTCGGTTCAGCGACACCAGGTTCCAGTACTCGTCGGTGCCACTACCTCAGTTGGCGAGCGGCGGGAAATGGTTCAATGAATTGCCACCGGAATTCTTTAGGAAAGTGTATAGAAAACCAAAGCCAGTCGATCCTGAAGTTTTGAAATCGATGAAAATGCAAGGTCCAATAGGATACGCGCCTAATCCGAAAACTTTTAAAAGGAATCAGATGCCGTACCTTGAAGAATCGTTAGAAGAAATGAGTATATCAAAAATAAATGGAAATAAAGTAATAATGCAGCCGATACCGAAACATTATCAGAAGCTCGACATACGATATAATAAGCACGGTCCGAATGAAGAATTAGAGAGTTTCAATAAAACAGGTCAGCCCGGACTGGAAGCGACATTACCGAATTCATACTGCAATGCCATGTTACAGGTTTTGTACTACACGCCGCCGGTCAAAGCCACGTTGCTAGCGCACACGTGCGCCAAAGAGTTCTGTTTGAGCTGCGAATTAGGTTTTCTATTTCGCATGCTGGACACGTCTGGCGGCACCCCTTGTCAGGCCACAAACTTCCTACGCGCGTTTCGTACGGTCCCCGAAGCGGCTGCCCTAGGACTGATACTGCCGGATCGAGCGCCCGACATAAGAGTCGATTTGGTAGCCTTAATACAGAGCTGGAACCGCTTCATCCTGCACCAGATTCATTACGAGATCCTCGAGACCCGTAAGAAGGAGAAGGAGTTAGCGCTGATAGCGAACAGTCCGCCCAAAGCACCACGCGCGTCCAACCCGAAGGAACGTCACTCGCTAATGAACGGACAGTACTCCGAGGAGTGTCAGTATACCGAACTGGAGTTCTTGGGACCCACCGAATTTGATTGTCAAGAGGATGGCGAACTCTGGCCGGGCAGGGATGAAGGCGTCGACGGCGTGGGGCCGTTGCAAAACCACGAAAGTCACGAGCTGAACAACCGACAGGCGCACCGGGAGGAGACCGAGATCTCGCAGCTGTTCGCCATCGGTCGGCATCAGCTCAACCGCTGCTTGAAGTGCAACAAGGAGGAGGGCAGGGAGTCGGTGGTGCTGGCGTGCGCCCTGCAGTACCCGCCGACGGGCGCGGGCGGCGCCGGGGAGGCGAGGGGCGGATTCGTCGAACTCGTGCGCGCGTCGCTGGCGGCGCGGCGGAGCACGCCGGCGTGGTGCGACCACTGCTCGCGGTTCACACCGACCACGCAGCGAGGGCGACTGCTGCGACTACCTCCTATATTGGCGATTAATTGTGGAGGAGTTACGGTCAGAGAGAAAGCGTATTGGGCTAAAGGAGTACAAAAGGAAATGTCCGATTCGAAAAGAGGGGGCAGCGGGAAACCGTGTCGCTACGGCTTACATTGCGCACGACCGGGTTGTCATTTCAAACATCCCGATCGGCCGAATACATCACAAAGTTCTTCCAAGGGCTCGAGCCAGGAGTCGCACTGCATGCTGCCGCTTCAGCTGCTCATCCGGCTTCAGTCAGACGGTGACGTCATCATCAACGACAAGATCGAAACTAATTCAGCGGAGAGTTCGCCGATCAACCGACCGGACACGAGCAGGCACAAAAAGAAGTACGTGACCACGCACCTCGAGGAGGAGTACTCGCTCTCGGCCGCCGTCGTCTGCGTCGAGGACAACCCTAAGAACTTGGTCGCCTACATACACACGGTCCGGGACGGCGAAACTCAATGGTTCATGTACAACGATCTATGTATCGTTCCGGTGAGTACGGACGAGGTGATACATTTCGGCGTGTGGTGGAAAACGCCTTGCGTGCTATTCTACACGGCGACCGGCGGCGGACAGTGCAACGGTGACGTCAGCTCATAG
Protein
MEYHYSEICMPSDHLTDYTPPFIIPTHEGEYEARNSVLVDGGERFGVSAVVFDKHEELIWMGNQGGHVTSYYGPNMQKYTSFQVHDTDEVRDIITVEKCIYVLTKTTLRHQLRRGIPKHTHKSPNMTDMQCLYQVNSTKLLMGGHQDKLIDLDLNKMVETIIPIQEEGCAVLRSGGSSHVACGSANGIVALRDMRAPTKAEHTFRAHTACLSDLDMQGDLLLTCGFTQAVGGVVAEPYIVVWDVRCLRGSNPGQGGWTIHTTSPPLLLHFLPAFSGRAVAASGDGHVSLLHVNAPEERQSIFKVETHSSQCNVMDVASTSQALAFGDQSGHLNLFSPTQNHEPVYNNFSRSTEFADPVEGLPYVRFSDTRFQYSSVPLPQLASGGKWFNELPPEFFRKVYRKPKPVDPEVLKSMKMQGPIGYAPNPKTFKRNQMPYLEESLEEMSISKINGNKVIMQPIPKHYQKLDIRYNKHGPNEELESFNKTGQPGLEATLPNSYCNAMLQVLYYTPPVKATLLAHTCAKEFCLSCELGFLFRMLDTSGGTPCQATNFLRAFRTVPEAAALGLILPDRAPDIRVDLVALIQSWNRFILHQIHYEILETRKKEKELALIANSPPKAPRASNPKERHSLMNGQYSEECQYTELEFLGPTEFDCQEDGELWPGRDEGVDGVGPLQNHESHELNNRQAHREETEISQLFAIGRHQLNRCLKCNKEEGRESVVLACALQYPPTGAGGAGEARGGFVELVRASLAARRSTPAWCDHCSRFTPTTQRGRLLRLPPILAINCGGVTVREKAYWAKGVQKEMSDSKRGGSGKPCRYGLHCARPGCHFKHPDRPNTSQSSSKGSSQESHCMLPLQLLIRLQSDGDVIINDKIETNSAESSPINRPDTSRHKKKYVTTHLEEEYSLSAAVVCVEDNPKNLVAYIHTVRDGETQWFMYNDLCIVPVSTDEVIHFGVWWKTPCVLFYTATGGGQCNGDVSS
Summary
Description
Catalytic subunit of the poly(A)-nuclease (PAN) deadenylation complex, one of two cytoplasmic mRNA deadenylases involved in general and miRNA-mediated mRNA turnover. PAN specifically shortens poly(A) tails of RNA and the activity is stimulated by poly(A)-binding protein (PABP). PAN deadenylation is followed by rapid degradation of the shortened mRNA tails by the CCR4-NOT complex. Deadenylated mRNAs are then degraded by two alternative mechanisms, namely exosome-mediated 3'-5' exonucleolytic degradation, or deadenlyation-dependent mRNA decaping and subsequent 5'-3' exonucleolytic degradation by XRN1.
Catalytic Activity
Exonucleolytic cleavage of poly(A) to 5'-AMP.
Cofactor
a divalent metal cation
Subunit
Forms a heterotrimer with an asymmetric homodimer of the regulatory subunit PAN3 to form the poly(A)-nuclease (PAN) deadenylation complex.
Similarity
Belongs to the peptidase C19 family. PAN2 subfamily.
Keywords
Complete proteome
Cytoplasm
Exonuclease
Hydrolase
Metal-binding
mRNA processing
Nuclease
Nucleus
Reference proteome
Feature
chain PAN2-PAN3 deadenylation complex catalytic subunit PAN2
Uniprot
A0A2H1X1K5
A0A2A4J6D2
A0A3S2M593
A0A194Q169
A0A0N0PCF7
A0A0L0CMD5
+ More
A0A1A9UZF6 A0A1A9ZQL9 A0A1B0FCG7 A0A0A1WKY4 A0A1A9X9Q1 A0A0A1X745 W8C0U8 A0A034VFC4 W8BE31 A0A034VB09 A0A0K8UJW9 A0A0K8UMD6 A0A026WRW3 B4HSB9 B4JVF5 T1PLY6 A0A0B4LEY4 A0A1I8MWA2 A1Z7K9 A0A1I8MWA8 B4P3E8 A0A0Q5WKR0 B3N839 A0A0J9TWR7 A0A0J9R8L1 B3MIA4 B4KTP8 B4LNP4 A0A0Q9W2X4 A0A3B0JKW5 A0A1I8Q0L5 A0A1I8Q0K1 Q28Y86 A0A2P8XF45 A0A0R3NNZ7 A0A1W4UJY0 A0A195D8A5 A0A1W4UZ14 A0A0L7R7H3 A0A2A3EJH6 A0A0P6J585 B4MIX2 A0A1W4UKF3 A0A088AEH3 A0A1W4UXN6 E2BV67 A0A195BCN5 A0A158NFR0 F4X1C0 A0A151WNW1 A0A1Y1MHS7 B0WEM6 A0A151JSW3 E1ZXV4 A0A182H522 A0A1Q3F7P3 A0A0M4EVE5 A0A0N0BDL8 A0A1B6ER40 A0A0K8VDY8 A0A1B6CJM8 A0A1W4WJ71 A0A1W4WV13 A0A154PQJ1 A0A195DP94 Q7QG93 D6WYL7 A0A182PBX3 A0A182YA82 A0A1B6MGP3 A0A182MD40 A0A182LMD0 A0A1B6M3L7 A0A182K478 A0A182W2E6 A0A182RR09 A0A084WLR5 A0A182T4B7 A0A182UZY4 A0A182HFK7 A0A182IN81 A0A182XG05 A0A0V0G6P6 A0A0P4VMQ2 A0A1J1IDM9 A0A182ULA7 N6U8E4 U4UGF8 A0A023F1S7 A0A224X6Y7
A0A1A9UZF6 A0A1A9ZQL9 A0A1B0FCG7 A0A0A1WKY4 A0A1A9X9Q1 A0A0A1X745 W8C0U8 A0A034VFC4 W8BE31 A0A034VB09 A0A0K8UJW9 A0A0K8UMD6 A0A026WRW3 B4HSB9 B4JVF5 T1PLY6 A0A0B4LEY4 A0A1I8MWA2 A1Z7K9 A0A1I8MWA8 B4P3E8 A0A0Q5WKR0 B3N839 A0A0J9TWR7 A0A0J9R8L1 B3MIA4 B4KTP8 B4LNP4 A0A0Q9W2X4 A0A3B0JKW5 A0A1I8Q0L5 A0A1I8Q0K1 Q28Y86 A0A2P8XF45 A0A0R3NNZ7 A0A1W4UJY0 A0A195D8A5 A0A1W4UZ14 A0A0L7R7H3 A0A2A3EJH6 A0A0P6J585 B4MIX2 A0A1W4UKF3 A0A088AEH3 A0A1W4UXN6 E2BV67 A0A195BCN5 A0A158NFR0 F4X1C0 A0A151WNW1 A0A1Y1MHS7 B0WEM6 A0A151JSW3 E1ZXV4 A0A182H522 A0A1Q3F7P3 A0A0M4EVE5 A0A0N0BDL8 A0A1B6ER40 A0A0K8VDY8 A0A1B6CJM8 A0A1W4WJ71 A0A1W4WV13 A0A154PQJ1 A0A195DP94 Q7QG93 D6WYL7 A0A182PBX3 A0A182YA82 A0A1B6MGP3 A0A182MD40 A0A182LMD0 A0A1B6M3L7 A0A182K478 A0A182W2E6 A0A182RR09 A0A084WLR5 A0A182T4B7 A0A182UZY4 A0A182HFK7 A0A182IN81 A0A182XG05 A0A0V0G6P6 A0A0P4VMQ2 A0A1J1IDM9 A0A182ULA7 N6U8E4 U4UGF8 A0A023F1S7 A0A224X6Y7
EC Number
3.1.13.4
Pubmed
26354079
26108605
25830018
24495485
25348373
24508170
+ More
30249741 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25315136 23932717 24880343 17550304 22936249 15632085 29403074 26999592 20798317 21347285 21719571 28004739 26483478 12364791 18362917 19820115 25244985 20966253 24438588 27129103 23537049 25474469
30249741 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25315136 23932717 24880343 17550304 22936249 15632085 29403074 26999592 20798317 21347285 21719571 28004739 26483478 12364791 18362917 19820115 25244985 20966253 24438588 27129103 23537049 25474469
EMBL
ODYU01012657
SOQ59086.1
NWSH01002801
PCG67511.1
RSAL01000030
RVE51698.1
+ More
KQ459580 KPI99301.1 KQ460571 KPJ13954.1 JRES01000195 KNC33426.1 CCAG010002922 GBXI01015224 JAC99067.1 GBXI01007163 JAD07129.1 GAMC01011296 JAB95259.1 GAKP01018452 JAC40500.1 GAMC01011297 JAB95258.1 GAKP01018451 JAC40501.1 GDHF01025501 GDHF01019213 GDHF01015207 GDHF01008943 JAI26813.1 JAI33101.1 JAI37107.1 JAI43371.1 GDHF01024784 GDHF01009308 JAI27530.1 JAI43006.1 KK107119 QOIP01000009 EZA58790.1 RLU18781.1 CH480816 EDW47014.1 CH916375 EDV98423.1 KA649144 KA649806 AFP64435.1 AE013599 AHN56017.1 BT031142 CM000157 EDW89421.1 CH954177 KQS70830.1 EDV59452.1 CM002911 KMY92380.1 KMY92381.1 CH902619 EDV35949.1 CH933808 EDW09631.1 CH940648 EDW60114.2 KRF79250.1 OUUW01000001 SPP73876.1 CM000071 EAL26079.1 PYGN01002372 PSN30631.1 KRT02814.1 KQ976750 KYN08674.1 KQ414640 KOC66794.1 KZ288230 PBC31624.1 GDUN01000655 JAN95264.1 CH963719 EDW72061.2 GL450824 EFN80359.1 KQ976514 KYM82318.1 ADTU01014572 GL888528 EGI59792.1 KQ982893 KYQ49554.1 GEZM01036261 JAV82847.1 DS231910 EDS45743.1 KQ982011 KYN30679.1 GL435132 EFN73985.1 JXUM01110650 JXUM01110651 KQ565431 KXJ70982.1 GFDL01011469 JAV23576.1 CP012524 ALC41582.1 KQ435855 KOX70612.1 GECZ01029337 JAS40432.1 GDHF01015155 JAI37159.1 GEDC01023612 JAS13686.1 KQ435039 KZC14159.1 KQ980662 KYN14666.1 AAAB01008838 EAA05834.4 KQ971363 EFA09006.2 GEBQ01004860 JAT35117.1 AXCM01001761 GEBQ01009455 JAT30522.1 ATLV01024270 KE525351 KFB51159.1 APCN01006362 GECL01002345 JAP03779.1 GDKW01000752 JAI55843.1 CVRI01000047 CRK98320.1 APGK01038985 KB740966 ENN76921.1 KB632186 ERL89691.1 GBBI01003287 JAC15425.1 GFTR01008209 JAW08217.1
KQ459580 KPI99301.1 KQ460571 KPJ13954.1 JRES01000195 KNC33426.1 CCAG010002922 GBXI01015224 JAC99067.1 GBXI01007163 JAD07129.1 GAMC01011296 JAB95259.1 GAKP01018452 JAC40500.1 GAMC01011297 JAB95258.1 GAKP01018451 JAC40501.1 GDHF01025501 GDHF01019213 GDHF01015207 GDHF01008943 JAI26813.1 JAI33101.1 JAI37107.1 JAI43371.1 GDHF01024784 GDHF01009308 JAI27530.1 JAI43006.1 KK107119 QOIP01000009 EZA58790.1 RLU18781.1 CH480816 EDW47014.1 CH916375 EDV98423.1 KA649144 KA649806 AFP64435.1 AE013599 AHN56017.1 BT031142 CM000157 EDW89421.1 CH954177 KQS70830.1 EDV59452.1 CM002911 KMY92380.1 KMY92381.1 CH902619 EDV35949.1 CH933808 EDW09631.1 CH940648 EDW60114.2 KRF79250.1 OUUW01000001 SPP73876.1 CM000071 EAL26079.1 PYGN01002372 PSN30631.1 KRT02814.1 KQ976750 KYN08674.1 KQ414640 KOC66794.1 KZ288230 PBC31624.1 GDUN01000655 JAN95264.1 CH963719 EDW72061.2 GL450824 EFN80359.1 KQ976514 KYM82318.1 ADTU01014572 GL888528 EGI59792.1 KQ982893 KYQ49554.1 GEZM01036261 JAV82847.1 DS231910 EDS45743.1 KQ982011 KYN30679.1 GL435132 EFN73985.1 JXUM01110650 JXUM01110651 KQ565431 KXJ70982.1 GFDL01011469 JAV23576.1 CP012524 ALC41582.1 KQ435855 KOX70612.1 GECZ01029337 JAS40432.1 GDHF01015155 JAI37159.1 GEDC01023612 JAS13686.1 KQ435039 KZC14159.1 KQ980662 KYN14666.1 AAAB01008838 EAA05834.4 KQ971363 EFA09006.2 GEBQ01004860 JAT35117.1 AXCM01001761 GEBQ01009455 JAT30522.1 ATLV01024270 KE525351 KFB51159.1 APCN01006362 GECL01002345 JAP03779.1 GDKW01000752 JAI55843.1 CVRI01000047 CRK98320.1 APGK01038985 KB740966 ENN76921.1 KB632186 ERL89691.1 GBBI01003287 JAC15425.1 GFTR01008209 JAW08217.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000053240
UP000037069
UP000078200
+ More
UP000092445 UP000092444 UP000092443 UP000053097 UP000279307 UP000001292 UP000001070 UP000000803 UP000095301 UP000002282 UP000008711 UP000007801 UP000009192 UP000008792 UP000268350 UP000095300 UP000001819 UP000245037 UP000192221 UP000078542 UP000053825 UP000242457 UP000007798 UP000005203 UP000008237 UP000078540 UP000005205 UP000007755 UP000075809 UP000002320 UP000078541 UP000000311 UP000069940 UP000249989 UP000092553 UP000053105 UP000192223 UP000076502 UP000078492 UP000007062 UP000007266 UP000075885 UP000076408 UP000075883 UP000075882 UP000075881 UP000075920 UP000075900 UP000030765 UP000075901 UP000075903 UP000075840 UP000075880 UP000076407 UP000183832 UP000075902 UP000019118 UP000030742
UP000092445 UP000092444 UP000092443 UP000053097 UP000279307 UP000001292 UP000001070 UP000000803 UP000095301 UP000002282 UP000008711 UP000007801 UP000009192 UP000008792 UP000268350 UP000095300 UP000001819 UP000245037 UP000192221 UP000078542 UP000053825 UP000242457 UP000007798 UP000005203 UP000008237 UP000078540 UP000005205 UP000007755 UP000075809 UP000002320 UP000078541 UP000000311 UP000069940 UP000249989 UP000092553 UP000053105 UP000192223 UP000076502 UP000078492 UP000007062 UP000007266 UP000075885 UP000076408 UP000075883 UP000075882 UP000075881 UP000075920 UP000075900 UP000030765 UP000075901 UP000075903 UP000075840 UP000075880 UP000076407 UP000183832 UP000075902 UP000019118 UP000030742
PRIDE
Interpro
IPR038765
Papain-like_cys_pep_sf
+ More
IPR036322 WD40_repeat_dom_sf
IPR028889 USP_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR028881 PAN2_dom
IPR001451 Hexapep
IPR011004 Trimer_LpxA-like_sf
IPR030843 PAN2
IPR013520 Exonuclease_RNaseT/DNA_pol3
IPR036397 RNaseH_sf
IPR012337 RNaseH-like_sf
IPR012423 Eaf7/MRGBP
IPR000571 Znf_CCCH
IPR011047 Quinoprotein_ADH-like_supfam
IPR036322 WD40_repeat_dom_sf
IPR028889 USP_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR028881 PAN2_dom
IPR001451 Hexapep
IPR011004 Trimer_LpxA-like_sf
IPR030843 PAN2
IPR013520 Exonuclease_RNaseT/DNA_pol3
IPR036397 RNaseH_sf
IPR012337 RNaseH-like_sf
IPR012423 Eaf7/MRGBP
IPR000571 Znf_CCCH
IPR011047 Quinoprotein_ADH-like_supfam
SUPFAM
Gene 3D
ProteinModelPortal
A0A2H1X1K5
A0A2A4J6D2
A0A3S2M593
A0A194Q169
A0A0N0PCF7
A0A0L0CMD5
+ More
A0A1A9UZF6 A0A1A9ZQL9 A0A1B0FCG7 A0A0A1WKY4 A0A1A9X9Q1 A0A0A1X745 W8C0U8 A0A034VFC4 W8BE31 A0A034VB09 A0A0K8UJW9 A0A0K8UMD6 A0A026WRW3 B4HSB9 B4JVF5 T1PLY6 A0A0B4LEY4 A0A1I8MWA2 A1Z7K9 A0A1I8MWA8 B4P3E8 A0A0Q5WKR0 B3N839 A0A0J9TWR7 A0A0J9R8L1 B3MIA4 B4KTP8 B4LNP4 A0A0Q9W2X4 A0A3B0JKW5 A0A1I8Q0L5 A0A1I8Q0K1 Q28Y86 A0A2P8XF45 A0A0R3NNZ7 A0A1W4UJY0 A0A195D8A5 A0A1W4UZ14 A0A0L7R7H3 A0A2A3EJH6 A0A0P6J585 B4MIX2 A0A1W4UKF3 A0A088AEH3 A0A1W4UXN6 E2BV67 A0A195BCN5 A0A158NFR0 F4X1C0 A0A151WNW1 A0A1Y1MHS7 B0WEM6 A0A151JSW3 E1ZXV4 A0A182H522 A0A1Q3F7P3 A0A0M4EVE5 A0A0N0BDL8 A0A1B6ER40 A0A0K8VDY8 A0A1B6CJM8 A0A1W4WJ71 A0A1W4WV13 A0A154PQJ1 A0A195DP94 Q7QG93 D6WYL7 A0A182PBX3 A0A182YA82 A0A1B6MGP3 A0A182MD40 A0A182LMD0 A0A1B6M3L7 A0A182K478 A0A182W2E6 A0A182RR09 A0A084WLR5 A0A182T4B7 A0A182UZY4 A0A182HFK7 A0A182IN81 A0A182XG05 A0A0V0G6P6 A0A0P4VMQ2 A0A1J1IDM9 A0A182ULA7 N6U8E4 U4UGF8 A0A023F1S7 A0A224X6Y7
A0A1A9UZF6 A0A1A9ZQL9 A0A1B0FCG7 A0A0A1WKY4 A0A1A9X9Q1 A0A0A1X745 W8C0U8 A0A034VFC4 W8BE31 A0A034VB09 A0A0K8UJW9 A0A0K8UMD6 A0A026WRW3 B4HSB9 B4JVF5 T1PLY6 A0A0B4LEY4 A0A1I8MWA2 A1Z7K9 A0A1I8MWA8 B4P3E8 A0A0Q5WKR0 B3N839 A0A0J9TWR7 A0A0J9R8L1 B3MIA4 B4KTP8 B4LNP4 A0A0Q9W2X4 A0A3B0JKW5 A0A1I8Q0L5 A0A1I8Q0K1 Q28Y86 A0A2P8XF45 A0A0R3NNZ7 A0A1W4UJY0 A0A195D8A5 A0A1W4UZ14 A0A0L7R7H3 A0A2A3EJH6 A0A0P6J585 B4MIX2 A0A1W4UKF3 A0A088AEH3 A0A1W4UXN6 E2BV67 A0A195BCN5 A0A158NFR0 F4X1C0 A0A151WNW1 A0A1Y1MHS7 B0WEM6 A0A151JSW3 E1ZXV4 A0A182H522 A0A1Q3F7P3 A0A0M4EVE5 A0A0N0BDL8 A0A1B6ER40 A0A0K8VDY8 A0A1B6CJM8 A0A1W4WJ71 A0A1W4WV13 A0A154PQJ1 A0A195DP94 Q7QG93 D6WYL7 A0A182PBX3 A0A182YA82 A0A1B6MGP3 A0A182MD40 A0A182LMD0 A0A1B6M3L7 A0A182K478 A0A182W2E6 A0A182RR09 A0A084WLR5 A0A182T4B7 A0A182UZY4 A0A182HFK7 A0A182IN81 A0A182XG05 A0A0V0G6P6 A0A0P4VMQ2 A0A1J1IDM9 A0A182ULA7 N6U8E4 U4UGF8 A0A023F1S7 A0A224X6Y7
PDB
4D0K
E-value=2.57102e-24,
Score=282
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Shuttles between nucleus and cytoplasm. With evidence from 1 publications.
P-body Shuttles between nucleus and cytoplasm. With evidence from 1 publications.
Nucleus Shuttles between nucleus and cytoplasm. With evidence from 1 publications.
P-body Shuttles between nucleus and cytoplasm. With evidence from 1 publications.
Nucleus Shuttles between nucleus and cytoplasm. With evidence from 1 publications.
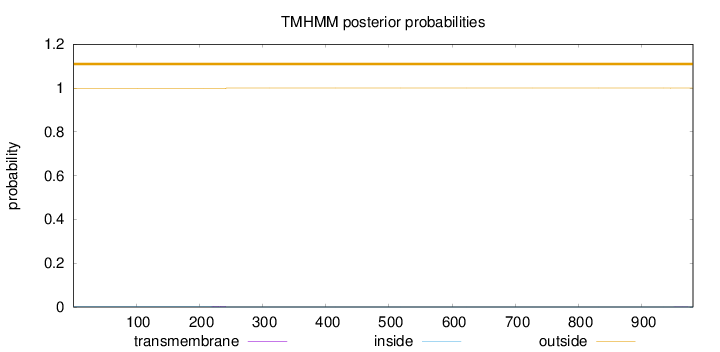
Length:
981
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0522400000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00199
outside
1 - 981
Population Genetic Test Statistics
Pi
229.062206
Theta
176.058956
Tajima's D
0.942018
CLR
0.329966
CSRT
0.640617969101545
Interpretation
Uncertain
