Gene
KWMTBOMO04378
Pre Gene Modal
BGIBMGA005273
Annotation
Uncharacterized_protein_OBRU01_10923_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 2.111
Sequence
CDS
ATGGAGTTTTGGGTTAAAAAAGTAGCTTTAATTACGGATTGTGGAGATGAATACGGAATGCATATAGCTGAAAAACTTAAGAAGTGCGGCATGACAGTTATTGGAATTGTTAAAAATGATAAGGAGACAAATATATTAGAAGAACAAATTATAGACAGACAAGGAATAGTGATATTTAAGTGCTGTAACATAAACATTGACAAATTCAATGAGATTTTTGAAAAAATCAATCATACTTACAAAGGAATCGATGTTATCATAAATAATTCGTATTGTAACATTAATACTAAAGTAAATGATGGAAAATTAACTGAAATTAAAAAGATAATTGATTTAAATATTAAACTAAATGTAGCAGCTACGAGACTCGCTGCACGATCAATGTTCGAAAGAAAATCTCGAGGCATTATTCTATTCATAGACAAGTTTAAGCATAGCAATAAAAAAGAAACTAAAGGTGTTTTCATAACTACAAAAGCAGCTATTGATGCGGTGAATGAAATTTTAAGAGTCGAATTGAAACATTTAAATGTGAACATTAAGGTAACAAATATATTATCACAGGCGAATTATGATAGACAAAATATTGCAACAAAAGAAGTTGATATGGAGGATGTTGCCGCAATGGTCGTTAAGGTTTTGGAAGCTCCTGAGAATCTACATATTCACGAAATATGGTTAGAAAAATCTGAATCTTGTTAA
Protein
MEFWVKKVALITDCGDEYGMHIAEKLKKCGMTVIGIVKNDKETNILEEQIIDRQGIVIFKCCNINIDKFNEIFEKINHTYKGIDVIINNSYCNINTKVNDGKLTEIKKIIDLNIKLNVAATRLAARSMFERKSRGIILFIDKFKHSNKKETKGVFITTKAAIDAVNEILRVELKHLNVNIKVTNILSQANYDRQNIATKEVDMEDVAAMVVKVLEAPENLHIHEIWLEKSESC
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
H9J6Y1
A0A2A4J8Z6
A0A2H1WZW0
A0A0L7LDQ5
A0A1I8NES6
A0A1A9WPM4
+ More
A0A0L0BP44 A0A349VJY2 A0A0L0BM36 A0A182PHM3 B4N2K2 A0A2E3X539 A0A1B0AX72 A0A1A9YNG7 A0A3M1TKR1 A0A182WII2 A0A2E8KLC4 A0A182Q801 A0A182K244 A0A182TGU9 A0A0P4WJ72 A0A1A9Z224 A0A1B0G4U0 A0A182RF95 A0A182MNA0 A0A3R7MET7 B4NV01 A0A182VHH0 A0A0P4W775 A0A182LJ42 Q7Q6V7 A0A182HWS6 A0A182XMX0 A0A182MQY1 A0A1I8MHY1 A0A1S3HIE8 A0A1W4WDZ6 B4R2S0 B4IDZ3 A0A1A9UUI4 A0A0L0BLT4 A0A1A9WPM3 A0A182YDX9 B4L3Z8 A0A182RS97 A0A1I8NQX2 A0A2P8ZKK7 Q9VZ19 C9QP79 Q16S89 A0A1S3H5Z0 B3NV94 Q8SY92 B3NY89 B3NY88 A0A0M3QZM0 A0A0L0BLQ8 A0A1A9YNG5 A0A3B3ZW86 A0A1I8N4V0 A0A1W4W217 B3MYJ4 A0A336MY33 A0A1I8MBG2 A0A1A9YNG4 A0A182W7X9 A0A1S3IMD5 A0A182T7E3 B4GXK4 A0A1I8Q6A2 A0A1I8MV30 A0A1S3HI38 A0A2M4BXZ0 Q7Q0J5 A0A1S4FRH3 D3TQS0 A0A2E8S1I2 A0A182XT50 A0A182IHL4 A0A182MFS5 B3MVY4 B4PYE3 Q29HJ6 A0A1W4X3Z7 B4PZM4 A0A182Y0X1 B3MYJ5 A0A1S3IAA7 A0A1A8LWS2 B4IKP3 A0A1I8NRH8 A0A0M3QZE9 Q5LJT3 A0A1I8N8F7 A0A1A9Z222 A0A1I8PF16 A0A1S3I357 A0A1A8NM55 A0A2J7QD23 A0A1B0AIK7
A0A0L0BP44 A0A349VJY2 A0A0L0BM36 A0A182PHM3 B4N2K2 A0A2E3X539 A0A1B0AX72 A0A1A9YNG7 A0A3M1TKR1 A0A182WII2 A0A2E8KLC4 A0A182Q801 A0A182K244 A0A182TGU9 A0A0P4WJ72 A0A1A9Z224 A0A1B0G4U0 A0A182RF95 A0A182MNA0 A0A3R7MET7 B4NV01 A0A182VHH0 A0A0P4W775 A0A182LJ42 Q7Q6V7 A0A182HWS6 A0A182XMX0 A0A182MQY1 A0A1I8MHY1 A0A1S3HIE8 A0A1W4WDZ6 B4R2S0 B4IDZ3 A0A1A9UUI4 A0A0L0BLT4 A0A1A9WPM3 A0A182YDX9 B4L3Z8 A0A182RS97 A0A1I8NQX2 A0A2P8ZKK7 Q9VZ19 C9QP79 Q16S89 A0A1S3H5Z0 B3NV94 Q8SY92 B3NY89 B3NY88 A0A0M3QZM0 A0A0L0BLQ8 A0A1A9YNG5 A0A3B3ZW86 A0A1I8N4V0 A0A1W4W217 B3MYJ4 A0A336MY33 A0A1I8MBG2 A0A1A9YNG4 A0A182W7X9 A0A1S3IMD5 A0A182T7E3 B4GXK4 A0A1I8Q6A2 A0A1I8MV30 A0A1S3HI38 A0A2M4BXZ0 Q7Q0J5 A0A1S4FRH3 D3TQS0 A0A2E8S1I2 A0A182XT50 A0A182IHL4 A0A182MFS5 B3MVY4 B4PYE3 Q29HJ6 A0A1W4X3Z7 B4PZM4 A0A182Y0X1 B3MYJ5 A0A1S3IAA7 A0A1A8LWS2 B4IKP3 A0A1I8NRH8 A0A0M3QZE9 Q5LJT3 A0A1I8N8F7 A0A1A9Z222 A0A1I8PF16 A0A1S3I357 A0A1A8NM55 A0A2J7QD23 A0A1B0AIK7
Pubmed
EMBL
BABH01028436
NWSH01002579
PCG67930.1
ODYU01012363
SOQ58640.1
JTDY01001623
+ More
KOB73321.1 JRES01001683 KNC20984.1 DMZP01000166 HAU48847.1 KNC20988.1 CH963925 EDW78591.1 PATN01000034 MBC89813.1 JXJN01005099 RFHS01000238 RMG17250.1 PBRR01000029 MBQ79183.1 AXCN02000361 GDRN01085557 JAI61334.1 CCAG010014629 AXCM01001467 QCYY01001185 ROT79845.1 CH985115 EDX16272.1 GDRN01080859 JAI62123.1 AAAB01008960 EAA11743.4 APCN01001535 AXCM01015925 CM000366 EDX17619.1 CH480830 EDW45801.1 KNC20987.1 CH933810 EDW07276.1 PYGN01000028 PSN57034.1 AE014298 AAF48012.1 BT099961 ACX47662.1 CH477680 EAT37326.1 CH954180 EDV46082.1 AY071705 AAL49327.1 EDV47674.1 EDV47673.1 CP012528 ALC49653.1 KNC20986.1 CH902632 EDV32688.1 UFQS01002357 UFQT01001486 UFQT01002357 SSX13864.1 SSX30768.1 CH479196 EDW27481.1 GGFJ01008805 MBW57946.1 AAAB01008981 EAA14615.3 EZ423772 ADD20048.1 PBVT01000081 MBR58437.1 APCN01007964 APCN01007965 AXCM01000973 CH902625 EDV35129.1 CM000162 EDX02005.1 CH379064 EAL31762.1 EDX03152.1 EDV32689.1 HAEF01009672 SBR48953.1 CH480854 EDW51657.1 ALC49303.1 BT081977 ACO53092.1 EAL24567.1 HAEH01002971 HAEI01007020 SBR70066.1 NEVH01015829 PNF26475.1
KOB73321.1 JRES01001683 KNC20984.1 DMZP01000166 HAU48847.1 KNC20988.1 CH963925 EDW78591.1 PATN01000034 MBC89813.1 JXJN01005099 RFHS01000238 RMG17250.1 PBRR01000029 MBQ79183.1 AXCN02000361 GDRN01085557 JAI61334.1 CCAG010014629 AXCM01001467 QCYY01001185 ROT79845.1 CH985115 EDX16272.1 GDRN01080859 JAI62123.1 AAAB01008960 EAA11743.4 APCN01001535 AXCM01015925 CM000366 EDX17619.1 CH480830 EDW45801.1 KNC20987.1 CH933810 EDW07276.1 PYGN01000028 PSN57034.1 AE014298 AAF48012.1 BT099961 ACX47662.1 CH477680 EAT37326.1 CH954180 EDV46082.1 AY071705 AAL49327.1 EDV47674.1 EDV47673.1 CP012528 ALC49653.1 KNC20986.1 CH902632 EDV32688.1 UFQS01002357 UFQT01001486 UFQT01002357 SSX13864.1 SSX30768.1 CH479196 EDW27481.1 GGFJ01008805 MBW57946.1 AAAB01008981 EAA14615.3 EZ423772 ADD20048.1 PBVT01000081 MBR58437.1 APCN01007964 APCN01007965 AXCM01000973 CH902625 EDV35129.1 CM000162 EDX02005.1 CH379064 EAL31762.1 EDX03152.1 EDV32689.1 HAEF01009672 SBR48953.1 CH480854 EDW51657.1 ALC49303.1 BT081977 ACO53092.1 EAL24567.1 HAEH01002971 HAEI01007020 SBR70066.1 NEVH01015829 PNF26475.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000095301
UP000091820
UP000037069
+ More
UP000075885 UP000007798 UP000092460 UP000092443 UP000075920 UP000075886 UP000075881 UP000075902 UP000092445 UP000092444 UP000075900 UP000075883 UP000283509 UP000000304 UP000075903 UP000075882 UP000007062 UP000075840 UP000076407 UP000085678 UP000192221 UP000001292 UP000078200 UP000076408 UP000009192 UP000095300 UP000245037 UP000000803 UP000008820 UP000008711 UP000092553 UP000261520 UP000007801 UP000075901 UP000008744 UP000002282 UP000001819 UP000192223 UP000235965
UP000075885 UP000007798 UP000092460 UP000092443 UP000075920 UP000075886 UP000075881 UP000075902 UP000092445 UP000092444 UP000075900 UP000075883 UP000283509 UP000000304 UP000075903 UP000075882 UP000007062 UP000075840 UP000076407 UP000085678 UP000192221 UP000001292 UP000078200 UP000076408 UP000009192 UP000095300 UP000245037 UP000000803 UP000008820 UP000008711 UP000092553 UP000261520 UP000007801 UP000075901 UP000008744 UP000002282 UP000001819 UP000192223 UP000235965
Pfam
PF00106 adh_short
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9J6Y1
A0A2A4J8Z6
A0A2H1WZW0
A0A0L7LDQ5
A0A1I8NES6
A0A1A9WPM4
+ More
A0A0L0BP44 A0A349VJY2 A0A0L0BM36 A0A182PHM3 B4N2K2 A0A2E3X539 A0A1B0AX72 A0A1A9YNG7 A0A3M1TKR1 A0A182WII2 A0A2E8KLC4 A0A182Q801 A0A182K244 A0A182TGU9 A0A0P4WJ72 A0A1A9Z224 A0A1B0G4U0 A0A182RF95 A0A182MNA0 A0A3R7MET7 B4NV01 A0A182VHH0 A0A0P4W775 A0A182LJ42 Q7Q6V7 A0A182HWS6 A0A182XMX0 A0A182MQY1 A0A1I8MHY1 A0A1S3HIE8 A0A1W4WDZ6 B4R2S0 B4IDZ3 A0A1A9UUI4 A0A0L0BLT4 A0A1A9WPM3 A0A182YDX9 B4L3Z8 A0A182RS97 A0A1I8NQX2 A0A2P8ZKK7 Q9VZ19 C9QP79 Q16S89 A0A1S3H5Z0 B3NV94 Q8SY92 B3NY89 B3NY88 A0A0M3QZM0 A0A0L0BLQ8 A0A1A9YNG5 A0A3B3ZW86 A0A1I8N4V0 A0A1W4W217 B3MYJ4 A0A336MY33 A0A1I8MBG2 A0A1A9YNG4 A0A182W7X9 A0A1S3IMD5 A0A182T7E3 B4GXK4 A0A1I8Q6A2 A0A1I8MV30 A0A1S3HI38 A0A2M4BXZ0 Q7Q0J5 A0A1S4FRH3 D3TQS0 A0A2E8S1I2 A0A182XT50 A0A182IHL4 A0A182MFS5 B3MVY4 B4PYE3 Q29HJ6 A0A1W4X3Z7 B4PZM4 A0A182Y0X1 B3MYJ5 A0A1S3IAA7 A0A1A8LWS2 B4IKP3 A0A1I8NRH8 A0A0M3QZE9 Q5LJT3 A0A1I8N8F7 A0A1A9Z222 A0A1I8PF16 A0A1S3I357 A0A1A8NM55 A0A2J7QD23 A0A1B0AIK7
A0A0L0BP44 A0A349VJY2 A0A0L0BM36 A0A182PHM3 B4N2K2 A0A2E3X539 A0A1B0AX72 A0A1A9YNG7 A0A3M1TKR1 A0A182WII2 A0A2E8KLC4 A0A182Q801 A0A182K244 A0A182TGU9 A0A0P4WJ72 A0A1A9Z224 A0A1B0G4U0 A0A182RF95 A0A182MNA0 A0A3R7MET7 B4NV01 A0A182VHH0 A0A0P4W775 A0A182LJ42 Q7Q6V7 A0A182HWS6 A0A182XMX0 A0A182MQY1 A0A1I8MHY1 A0A1S3HIE8 A0A1W4WDZ6 B4R2S0 B4IDZ3 A0A1A9UUI4 A0A0L0BLT4 A0A1A9WPM3 A0A182YDX9 B4L3Z8 A0A182RS97 A0A1I8NQX2 A0A2P8ZKK7 Q9VZ19 C9QP79 Q16S89 A0A1S3H5Z0 B3NV94 Q8SY92 B3NY89 B3NY88 A0A0M3QZM0 A0A0L0BLQ8 A0A1A9YNG5 A0A3B3ZW86 A0A1I8N4V0 A0A1W4W217 B3MYJ4 A0A336MY33 A0A1I8MBG2 A0A1A9YNG4 A0A182W7X9 A0A1S3IMD5 A0A182T7E3 B4GXK4 A0A1I8Q6A2 A0A1I8MV30 A0A1S3HI38 A0A2M4BXZ0 Q7Q0J5 A0A1S4FRH3 D3TQS0 A0A2E8S1I2 A0A182XT50 A0A182IHL4 A0A182MFS5 B3MVY4 B4PYE3 Q29HJ6 A0A1W4X3Z7 B4PZM4 A0A182Y0X1 B3MYJ5 A0A1S3IAA7 A0A1A8LWS2 B4IKP3 A0A1I8NRH8 A0A0M3QZE9 Q5LJT3 A0A1I8N8F7 A0A1A9Z222 A0A1I8PF16 A0A1S3I357 A0A1A8NM55 A0A2J7QD23 A0A1B0AIK7
PDB
1XG5
E-value=4.65046e-07,
Score=126
Ontologies
KEGG
GO
Topology
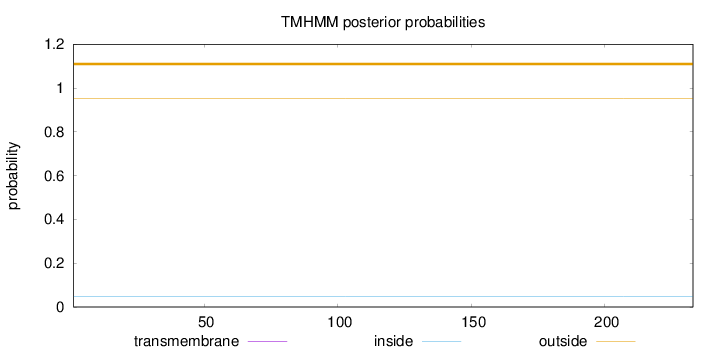
Length:
233
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000550000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04640
outside
1 - 233
Population Genetic Test Statistics
Pi
151.079709
Theta
150.617599
Tajima's D
0.009645
CLR
0
CSRT
0.371431428428579
Interpretation
Uncertain
