Gene
KWMTBOMO04351
Pre Gene Modal
BGIBMGA000832
Annotation
PREDICTED:_fibroblast_growth_factor_receptor_2-like_isoform_X1_[Amyelois_transitella]
Full name
Flap endonuclease 1
Alternative Name
Flap structure-specific endonuclease 1
Location in the cell
Cytoplasmic Reliability : 1.977 Nuclear Reliability : 1.335
Sequence
CDS
ATGAGTACCAAAAACACAATTATTATAATCAAAGAAGAAGCATCGCAGGAAGAGATGCAAGACTTCTTCCGGGAAATAGAGATGCTGAAGCACGTTGGCTACCACAAACACGTGATTCGTCTCATCGGCTGCTGCACGAGACGAGCCCCGCTCATCGCTCTCCTGGAACACGCTCCAAGAGGTGATCTGCTTTCACTGTTGAGGGCAGCCAGAGGGAGAAGGAAGGAAACTCAGACGTCTGACGTCACTAGGAGAGCAGGCAGCGACATTATCGGAAAACCTTCTGAAGGGGATTCGGATTACACACATTTTAGCGACTCGGATCCGGCACTGTCGGACGAAAAACTTTACATAGACAGCGAAATAAATAAATTAATGAAAAGAGACCATTACGTGGCTGAGCCTGCGTTACATTTAGATAGTACGACTATGAGAGAATACGCGTTGCAAGTCGCTTTAGGAATGAAGCATTTGGAGGGCAGAGGGATCACGCATAGAGATCTGGCGGCTCGGAACATCCTGGTGGATAGCACGGGAGTCTTGAAAGTGGCAGACTTCGGCCTCTCCAGATCAGGAGTGTACGTCCACACGCGTTCCAAGCCAGTGCCGCTGCGATGGCTAGCACCCGAAGCCATTGTCCATTCCCAATATTGCAGCAAAAGCGATGTTTGGGCTTTCGCTGTTTTGCTTTGGGAGATAGCCACCTTGGGTGGTTTCCCTTACGCGGAACTAAGCAACTACCAAGTACCCGCATTTTTGACTGGCGGCGGCAGACTGCCGAAGCCAATGCGAGCGTCAGTGCGCCTCTACGAATTAATGGTCGAATGTTGGTCAGACGATCCACACGACCGGCCAACGTTCGCTCAAATAGTGGACAAACTGGTCATCCAACAGCAGTTGTATGTTGATTTGGAGTGCGTGCTTCCACCGTCAGAAGAAGACATTGGATTCAAAGATTATGATTACACTCTGCCAAGCCCACTCCCCGGGGACACCCAGCTTATGTACAAAGACTGA
Protein
MSTKNTIIIIKEEASQEEMQDFFREIEMLKHVGYHKHVIRLIGCCTRRAPLIALLEHAPRGDLLSLLRAARGRRKETQTSDVTRRAGSDIIGKPSEGDSDYTHFSDSDPALSDEKLYIDSEINKLMKRDHYVAEPALHLDSTTMREYALQVALGMKHLEGRGITHRDLAARNILVDSTGVLKVADFGLSRSGVYVHTRSKPVPLRWLAPEAIVHSQYCSKSDVWAFAVLLWEIATLGGFPYAELSNYQVPAFLTGGGRLPKPMRASVRLYELMVECWSDDPHDRPTFAQIVDKLVIQQQLYVDLECVLPPSEEDIGFKDYDYTLPSPLPGDTQLMYKD
Summary
Description
Structure-specific nuclease with 5'-flap endonuclease and 5'-3' exonuclease activities involved in DNA replication and repair. During DNA replication, cleaves the 5'-overhanging flap structure that is generated by displacement synthesis when DNA polymerase encounters the 5'-end of a downstream Okazaki fragment. It enters the flap from the 5'-end and then tracks to cleave the flap base, leaving a nick for ligation. Also involved in the long patch base excision repair (LP-BER) pathway, by cleaving within the apurinic/apyrimidinic (AP) site-terminated flap. Acts as a genome stabilization factor that prevents flaps from equilibrating into structurs that lead to duplications and deletions. Also possesses 5'-3' exonuclease activity on nicked or gapped double-stranded DNA, and exhibits RNase H activity. Also involved in replication and repair of rDNA and in repairing mitochondrial DNA.
Catalytic Activity
ATP + L-tyrosyl-[protein] = ADP + H(+) + O-phospho-L-tyrosyl-[protein]
Cofactor
Mg(2+)
Subunit
Interacts with PCNA. Three molecules of sls bind to one PCNA trimer with each molecule binding to one PCNA monomer. PCNA stimulates the nuclease activity without altering cleavage specificity.
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Belongs to the XPG/RAD2 endonuclease family. FEN1 subfamily.
Belongs to the XPG/RAD2 endonuclease family. FEN1 subfamily.
Uniprot
A0A2H1WPD8
A0A2A4JIF0
A0A194Q2T8
A0A0N1IAD9
A0A3S2PHM8
A0A212EH28
+ More
A0A0L7KY07 A0A182UEF3 A0A1S4H435 A0A1B6DW38 A0A1B6CL53 A0A084VPY9 A0A2J7RRJ4 A0A182FMY7 A0A182Q8T9 Q179Z3 A0A1L8DJG6 A0A1L8DJG5 A0A182JN47 A0A182N8P0 W5J967 Q7Q0U2 V5IAC6 A0A067R3X6 A0A1B6MNC8 A0A182KJM7 A0A1B6LTA0 A0A0L0C7C2 A0A182PVI5 A0A139WC45 B3NAS7 B4Q955 B4I2X7 B4NXU5 A0A1B6IHA6 W8BCC2 A0A3P7E412 W8BST5 Q8IQ00 A0A1I8E8P7 A0A0N4U1K1 A0A0N5DU80 B4NLR3 A0A0J9XQ09 A0A1I7V8S3 A0A3P6TXP8 A0A1S0UJ96 A0A077Z082 A0A0A1XQY7 B3MNY1 A0A0M4ECA8 A0A044UX42 A0A1W4UU21 A0A158RBL0 A0A034VDX9 A0A085NNF4 B4JDX2 B4LTH2 A0A085MGX7 F1KW71 A0A0K8UN28 A0A0A1XCK9 A0A183V2U5 B4G9R0 Q29M57 A0A0R3NY09 A0A1Y1L060 A0A158R5H6 A0A0V1D1Q4 A0A0V1HJV2 A0A0V1HHN3 A0A0V1HIA9 A0A0V1HHS0 A0A0V1HJ59 A0A0V1HHZ4 A0A0V1HIK5 A0A0V1N404 A0A0V1HI03 A0A0V1HJ22 A0A0V1HIJ2 A0A0V0ZYW2 A0A0V0ZYN5 A0A0V0ZYT7
A0A0L7KY07 A0A182UEF3 A0A1S4H435 A0A1B6DW38 A0A1B6CL53 A0A084VPY9 A0A2J7RRJ4 A0A182FMY7 A0A182Q8T9 Q179Z3 A0A1L8DJG6 A0A1L8DJG5 A0A182JN47 A0A182N8P0 W5J967 Q7Q0U2 V5IAC6 A0A067R3X6 A0A1B6MNC8 A0A182KJM7 A0A1B6LTA0 A0A0L0C7C2 A0A182PVI5 A0A139WC45 B3NAS7 B4Q955 B4I2X7 B4NXU5 A0A1B6IHA6 W8BCC2 A0A3P7E412 W8BST5 Q8IQ00 A0A1I8E8P7 A0A0N4U1K1 A0A0N5DU80 B4NLR3 A0A0J9XQ09 A0A1I7V8S3 A0A3P6TXP8 A0A1S0UJ96 A0A077Z082 A0A0A1XQY7 B3MNY1 A0A0M4ECA8 A0A044UX42 A0A1W4UU21 A0A158RBL0 A0A034VDX9 A0A085NNF4 B4JDX2 B4LTH2 A0A085MGX7 F1KW71 A0A0K8UN28 A0A0A1XCK9 A0A183V2U5 B4G9R0 Q29M57 A0A0R3NY09 A0A1Y1L060 A0A158R5H6 A0A0V1D1Q4 A0A0V1HJV2 A0A0V1HHN3 A0A0V1HIA9 A0A0V1HHS0 A0A0V1HJ59 A0A0V1HHZ4 A0A0V1HIK5 A0A0V1N404 A0A0V1HI03 A0A0V1HJ22 A0A0V1HIJ2 A0A0V0ZYW2 A0A0V0ZYN5 A0A0V0ZYT7
EC Number
3.1.-.-
Pubmed
26354079
22118469
26227816
12364791
24438588
17510324
+ More
20920257 23761445 24845553 20966253 26108605 18362917 19820115 17994087 22936249 17550304 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26850696 17885136 25217238 25830018 25348373 24929829 21685128 15632085 28004739
20920257 23761445 24845553 20966253 26108605 18362917 19820115 17994087 22936249 17550304 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26850696 17885136 25217238 25830018 25348373 24929829 21685128 15632085 28004739
EMBL
ODYU01010083
SOQ54935.1
NWSH01001362
PCG71546.1
KQ459580
KPI99314.1
+ More
KQ460393 KPJ15389.1 RSAL01000030 RVE51708.1 AGBW02014980 OWR40784.1 JTDY01004618 KOB67936.1 AAAB01008980 GEDC01007403 JAS29895.1 GEDC01023203 JAS14095.1 ATLV01015084 KE525002 KFB40033.1 NEVH01000606 PNF43435.1 AXCN02001858 CH477342 EAT43078.1 GFDF01007491 JAV06593.1 GFDF01007492 JAV06592.1 ADMH02002025 ETN59928.1 EAA13998.4 GALX01001420 JAB67046.1 KK852776 KDR16790.1 GEBQ01002509 JAT37468.1 GEBQ01013065 JAT26912.1 JRES01000799 KNC28293.1 KQ971371 KYB25529.1 CH954177 EDV57600.1 CM000361 CM002910 EDX03594.1 KMY87853.1 CH480820 EDW54122.1 CM000157 EDW87516.2 GECU01021405 JAS86301.1 GAMC01010213 JAB96342.1 UYWW01001541 VDM10659.1 GAMC01010214 JAB96341.1 AE014134 AAN10388.3 UYYG01001151 VDN54876.1 CH964274 EDW85302.1 LN856867 CDP93291.1 UYRX01001095 VDK88149.1 JH712198 EJD74947.1 HG805868 CDW53822.1 GBXI01000568 JAD13724.1 CH902620 EDV32168.2 CP012523 ALC39235.1 CMVM020000238 UYYF01004311 VDN02111.1 GAKP01019216 GAKP01019214 JAC39738.1 KL367484 KFD71000.1 CH916368 EDW03492.1 CH940649 EDW63942.2 KL363193 KFD56473.1 JI166901 ADY42125.1 GDHF01024584 JAI27730.1 GBXI01006094 JAD08198.1 UYWY01022611 VDM46386.1 CH479180 EDW29090.1 CH379060 EAL33838.3 KRT04255.1 GEZM01072991 JAV65335.1 JYDI01000055 KRY55434.1 JYDP01000061 KRZ10322.1 KRZ10318.1 KRZ10314.1 KRZ10315.1 KRZ10320.1 KRZ10317.1 KRZ10316.1 JYDO01000010 KRZ78775.1 KRZ10323.1 KRZ10319.1 KRZ10321.1 JYDQ01000055 KRY17832.1 KRY17830.1 KRY17831.1
KQ460393 KPJ15389.1 RSAL01000030 RVE51708.1 AGBW02014980 OWR40784.1 JTDY01004618 KOB67936.1 AAAB01008980 GEDC01007403 JAS29895.1 GEDC01023203 JAS14095.1 ATLV01015084 KE525002 KFB40033.1 NEVH01000606 PNF43435.1 AXCN02001858 CH477342 EAT43078.1 GFDF01007491 JAV06593.1 GFDF01007492 JAV06592.1 ADMH02002025 ETN59928.1 EAA13998.4 GALX01001420 JAB67046.1 KK852776 KDR16790.1 GEBQ01002509 JAT37468.1 GEBQ01013065 JAT26912.1 JRES01000799 KNC28293.1 KQ971371 KYB25529.1 CH954177 EDV57600.1 CM000361 CM002910 EDX03594.1 KMY87853.1 CH480820 EDW54122.1 CM000157 EDW87516.2 GECU01021405 JAS86301.1 GAMC01010213 JAB96342.1 UYWW01001541 VDM10659.1 GAMC01010214 JAB96341.1 AE014134 AAN10388.3 UYYG01001151 VDN54876.1 CH964274 EDW85302.1 LN856867 CDP93291.1 UYRX01001095 VDK88149.1 JH712198 EJD74947.1 HG805868 CDW53822.1 GBXI01000568 JAD13724.1 CH902620 EDV32168.2 CP012523 ALC39235.1 CMVM020000238 UYYF01004311 VDN02111.1 GAKP01019216 GAKP01019214 JAC39738.1 KL367484 KFD71000.1 CH916368 EDW03492.1 CH940649 EDW63942.2 KL363193 KFD56473.1 JI166901 ADY42125.1 GDHF01024584 JAI27730.1 GBXI01006094 JAD08198.1 UYWY01022611 VDM46386.1 CH479180 EDW29090.1 CH379060 EAL33838.3 KRT04255.1 GEZM01072991 JAV65335.1 JYDI01000055 KRY55434.1 JYDP01000061 KRZ10322.1 KRZ10318.1 KRZ10314.1 KRZ10315.1 KRZ10320.1 KRZ10317.1 KRZ10316.1 JYDO01000010 KRZ78775.1 KRZ10323.1 KRZ10319.1 KRZ10321.1 JYDQ01000055 KRY17832.1 KRY17830.1 KRY17831.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000283053
UP000007151
UP000037510
+ More
UP000075902 UP000030765 UP000235965 UP000069272 UP000075886 UP000008820 UP000075880 UP000075884 UP000000673 UP000007062 UP000027135 UP000075882 UP000037069 UP000075885 UP000007266 UP000008711 UP000000304 UP000001292 UP000002282 UP000270924 UP000000803 UP000093561 UP000038040 UP000274756 UP000046395 UP000007798 UP000006672 UP000095285 UP000277928 UP000030665 UP000007801 UP000092553 UP000024404 UP000192221 UP000046394 UP000276776 UP000001070 UP000008792 UP000050794 UP000267007 UP000008744 UP000001819 UP000046393 UP000054653 UP000055024 UP000054843 UP000054783
UP000075902 UP000030765 UP000235965 UP000069272 UP000075886 UP000008820 UP000075880 UP000075884 UP000000673 UP000007062 UP000027135 UP000075882 UP000037069 UP000075885 UP000007266 UP000008711 UP000000304 UP000001292 UP000002282 UP000270924 UP000000803 UP000093561 UP000038040 UP000274756 UP000046395 UP000007798 UP000006672 UP000095285 UP000277928 UP000030665 UP000007801 UP000092553 UP000024404 UP000192221 UP000046394 UP000276776 UP000001070 UP000008792 UP000050794 UP000267007 UP000008744 UP000001819 UP000046393 UP000054653 UP000055024 UP000054843 UP000054783
PRIDE
Pfam
Interpro
IPR008266
Tyr_kinase_AS
+ More
IPR020635 Tyr_kinase_cat_dom
IPR000719 Prot_kinase_dom
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR020683 Ankyrin_rpt-contain_dom
IPR021939 KN_motif
IPR036770 Ankyrin_rpt-contain_sf
IPR002110 Ankyrin_rpt
IPR013783 Ig-like_fold
IPR036279 5-3_exonuclease_C_sf
IPR024969 Rpn11/EIF3F_C
IPR029060 PIN-like_dom_sf
IPR006086 XPG-I_dom
IPR003961 FN3_dom
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR013098 Ig_I-set
IPR019974 XPG_CS
IPR008271 Ser/Thr_kinase_AS
IPR036116 FN3_sf
IPR006084 XPG/Rad2
IPR008918 HhH2
IPR000555 JAMM/MPN+_dom
IPR013106 Ig_V-set
IPR010939 Titin-rpts
IPR037518 MPN
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR006085 XPG_DNA_repair_N
IPR023426 Flap_endonuc
IPR018159 Spectrin/alpha-actinin
IPR002017 Spectrin_repeat
IPR020635 Tyr_kinase_cat_dom
IPR000719 Prot_kinase_dom
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR020683 Ankyrin_rpt-contain_dom
IPR021939 KN_motif
IPR036770 Ankyrin_rpt-contain_sf
IPR002110 Ankyrin_rpt
IPR013783 Ig-like_fold
IPR036279 5-3_exonuclease_C_sf
IPR024969 Rpn11/EIF3F_C
IPR029060 PIN-like_dom_sf
IPR006086 XPG-I_dom
IPR003961 FN3_dom
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR013098 Ig_I-set
IPR019974 XPG_CS
IPR008271 Ser/Thr_kinase_AS
IPR036116 FN3_sf
IPR006084 XPG/Rad2
IPR008918 HhH2
IPR000555 JAMM/MPN+_dom
IPR013106 Ig_V-set
IPR010939 Titin-rpts
IPR037518 MPN
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR006085 XPG_DNA_repair_N
IPR023426 Flap_endonuc
IPR018159 Spectrin/alpha-actinin
IPR002017 Spectrin_repeat
SUPFAM
Gene 3D
ProteinModelPortal
A0A2H1WPD8
A0A2A4JIF0
A0A194Q2T8
A0A0N1IAD9
A0A3S2PHM8
A0A212EH28
+ More
A0A0L7KY07 A0A182UEF3 A0A1S4H435 A0A1B6DW38 A0A1B6CL53 A0A084VPY9 A0A2J7RRJ4 A0A182FMY7 A0A182Q8T9 Q179Z3 A0A1L8DJG6 A0A1L8DJG5 A0A182JN47 A0A182N8P0 W5J967 Q7Q0U2 V5IAC6 A0A067R3X6 A0A1B6MNC8 A0A182KJM7 A0A1B6LTA0 A0A0L0C7C2 A0A182PVI5 A0A139WC45 B3NAS7 B4Q955 B4I2X7 B4NXU5 A0A1B6IHA6 W8BCC2 A0A3P7E412 W8BST5 Q8IQ00 A0A1I8E8P7 A0A0N4U1K1 A0A0N5DU80 B4NLR3 A0A0J9XQ09 A0A1I7V8S3 A0A3P6TXP8 A0A1S0UJ96 A0A077Z082 A0A0A1XQY7 B3MNY1 A0A0M4ECA8 A0A044UX42 A0A1W4UU21 A0A158RBL0 A0A034VDX9 A0A085NNF4 B4JDX2 B4LTH2 A0A085MGX7 F1KW71 A0A0K8UN28 A0A0A1XCK9 A0A183V2U5 B4G9R0 Q29M57 A0A0R3NY09 A0A1Y1L060 A0A158R5H6 A0A0V1D1Q4 A0A0V1HJV2 A0A0V1HHN3 A0A0V1HIA9 A0A0V1HHS0 A0A0V1HJ59 A0A0V1HHZ4 A0A0V1HIK5 A0A0V1N404 A0A0V1HI03 A0A0V1HJ22 A0A0V1HIJ2 A0A0V0ZYW2 A0A0V0ZYN5 A0A0V0ZYT7
A0A0L7KY07 A0A182UEF3 A0A1S4H435 A0A1B6DW38 A0A1B6CL53 A0A084VPY9 A0A2J7RRJ4 A0A182FMY7 A0A182Q8T9 Q179Z3 A0A1L8DJG6 A0A1L8DJG5 A0A182JN47 A0A182N8P0 W5J967 Q7Q0U2 V5IAC6 A0A067R3X6 A0A1B6MNC8 A0A182KJM7 A0A1B6LTA0 A0A0L0C7C2 A0A182PVI5 A0A139WC45 B3NAS7 B4Q955 B4I2X7 B4NXU5 A0A1B6IHA6 W8BCC2 A0A3P7E412 W8BST5 Q8IQ00 A0A1I8E8P7 A0A0N4U1K1 A0A0N5DU80 B4NLR3 A0A0J9XQ09 A0A1I7V8S3 A0A3P6TXP8 A0A1S0UJ96 A0A077Z082 A0A0A1XQY7 B3MNY1 A0A0M4ECA8 A0A044UX42 A0A1W4UU21 A0A158RBL0 A0A034VDX9 A0A085NNF4 B4JDX2 B4LTH2 A0A085MGX7 F1KW71 A0A0K8UN28 A0A0A1XCK9 A0A183V2U5 B4G9R0 Q29M57 A0A0R3NY09 A0A1Y1L060 A0A158R5H6 A0A0V1D1Q4 A0A0V1HJV2 A0A0V1HHN3 A0A0V1HIA9 A0A0V1HHS0 A0A0V1HJ59 A0A0V1HHZ4 A0A0V1HIK5 A0A0V1N404 A0A0V1HI03 A0A0V1HJ22 A0A0V1HIJ2 A0A0V0ZYW2 A0A0V0ZYN5 A0A0V0ZYT7
PDB
1FVR
E-value=1.43697e-40,
Score=417
Ontologies
GO
Topology
Subcellular location
Mitochondrion
Resides mostly in the nucleoli and relocalizes to the nucleoplasm upon DNA damage. With evidence from 1 publications.
Nucleus Resides mostly in the nucleoli and relocalizes to the nucleoplasm upon DNA damage. With evidence from 1 publications.
Nucleolus Resides mostly in the nucleoli and relocalizes to the nucleoplasm upon DNA damage. With evidence from 1 publications.
Nucleoplasm Resides mostly in the nucleoli and relocalizes to the nucleoplasm upon DNA damage. With evidence from 1 publications.
Nucleus Resides mostly in the nucleoli and relocalizes to the nucleoplasm upon DNA damage. With evidence from 1 publications.
Nucleolus Resides mostly in the nucleoli and relocalizes to the nucleoplasm upon DNA damage. With evidence from 1 publications.
Nucleoplasm Resides mostly in the nucleoli and relocalizes to the nucleoplasm upon DNA damage. With evidence from 1 publications.
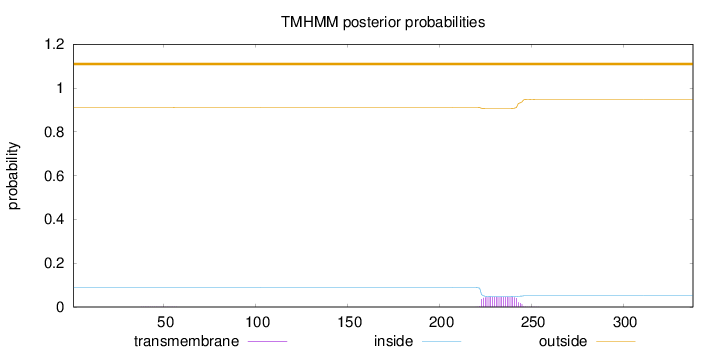
Length:
338
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.9646
Exp number, first 60 AAs:
0.00436
Total prob of N-in:
0.08924
outside
1 - 338
Population Genetic Test Statistics
Pi
216.538379
Theta
155.856448
Tajima's D
2.522032
CLR
0.358199
CSRT
0.946102694865257
Interpretation
Uncertain
