Gene
KWMTBOMO04347
Annotation
PREDICTED:_4-hydroxyphenylpyruvate_dioxygenase-like_[Amyelois_transitella]
Full name
4-hydroxyphenylpyruvate dioxygenase
Alternative Name
4-hydroxyphenylpyruvic acid oxidase
Location in the cell
Cytoplasmic Reliability : 1.6
Sequence
CDS
ATGCCTATAACCATAATATTCGAGTCACCCACAAAACCGGACAGTGAAATCGCGAGCAGCTTAAACACTCACGGAGACTTCGTGAAGGATGTGGCATTTGAAGTTCACAATCTCGACGAGCTGGTTCGGAGGGCCATAGAAGGGCGAGCCGAGATAATTAAAACAACAACACAAATTGAAGACAACGATGGTGTCTTGAAATACGCCATTTTGAAAACGTATGGAGAGAACACACATACGTTAATTGATCGGAGCAGATATAACGGTATCTTGTTGCCTGGATTCGTAGCCGTTGAAGATGATCCACTGACAGCGATGTTGTGA
Protein
MPITIIFESPTKPDSEIASSLNTHGDFVKDVAFEVHNLDELVRRAIEGRAEIIKTTTQIEDNDGVLKYAILKTYGENTHTLIDRSRYNGILLPGFVAVEDDPLTAML
Summary
Description
Key enzyme in the degradation of tyrosine.
Catalytic Activity
3-(4-hydroxyphenyl)pyruvate + O2 = CO2 + homogentisate
Cofactor
Fe cation
Similarity
Belongs to the 4HPPD family.
Keywords
Complete proteome
Dioxygenase
Iron
Metal-binding
Oxidoreductase
Phenylalanine catabolism
Reference proteome
Repeat
Tyrosine catabolism
Feature
chain 4-hydroxyphenylpyruvate dioxygenase
Uniprot
A0A2A4IY11
A0A2A4IUV6
A0A194Q7D9
A0A194QLP7
A0A0L7KYG5
A0A212EXC3
+ More
A0A212EYA1 A0A194QMV1 A0A2A4ISS0 I4DJI5 A0A194Q189 A0A2A4IS65 A0A195FM03 A0A3L8DEL4 H9J6X0 A0A140DKW1 A0A1I8IJD7 F4WEM2 E2BIZ1 A0A210PRQ4 A0A195C642 E9IWI6 A0A2H1W9R5 A0A0K8TWD3 A0A1S3JV90 A0A026WR60 A0A158NF97 A0A2A3EPS1 A0A0C9RMG1 A0A195AVG6 A0A088ACK9 A0A0N0U440 A0A0N4XB01 A0A267E053 A0A267H602 A0A1W4WLA9 A0A232F978 A0A1I8IMR3 F1L6R5 A0A2H2IKY3 A0A2B7Y825 A0A1D1VAW0 A0A165JYY0 A0A2C9JS97 D6WZP4 A0A0P6ENL0 A0A0B1TK92 A0A0P6DRR7 A0A0P5VJ77 A0A0U2N329 A0A0P6DF79 A0A0P5F559 K7J3E2 A0A0P5VD00 W6NC52 A0A0P5AQJ9 A0A0P5BJ32 A0A0P5NZL2 A0A0P5N2X2 A0A0P5ATU2 A0A0P5XAL6 Q22633 A0A0N8A5M5 A0A0P5BLJ0 A0A0P5MUW4 A0A0P5Q3X8 A0A0P4ZGJ1 A0A0P5QA72 A0A3B5A4L3 A0A151WHV6 W8B5L7 W8AU74 T1FNN4 R7V6N7 A0A3Q1J7S4 E2AE27 A0A3P8Z521 A0A3S3NZJ3 A0A3B3QJX6 A0A1I7TMB3 A0A0P5GQT2 A0A2G9TNT5 A0A2P8XHR9 A0A0P5SQ41 A0A2P4VGT7 A0A261BV92 E3LZD7 A0A3P8Z5R8 A0A0P6B140 A0A395GUJ1 V4A0B7 A0A3N0YSI0 A0A3P8Z6J3 A0A2G9UN36 Q6AZV8 F1Q986 A0A1B6M4D0 A0A060WF31 A0A0L7QZ05 A0A1S3PI18
A0A212EYA1 A0A194QMV1 A0A2A4ISS0 I4DJI5 A0A194Q189 A0A2A4IS65 A0A195FM03 A0A3L8DEL4 H9J6X0 A0A140DKW1 A0A1I8IJD7 F4WEM2 E2BIZ1 A0A210PRQ4 A0A195C642 E9IWI6 A0A2H1W9R5 A0A0K8TWD3 A0A1S3JV90 A0A026WR60 A0A158NF97 A0A2A3EPS1 A0A0C9RMG1 A0A195AVG6 A0A088ACK9 A0A0N0U440 A0A0N4XB01 A0A267E053 A0A267H602 A0A1W4WLA9 A0A232F978 A0A1I8IMR3 F1L6R5 A0A2H2IKY3 A0A2B7Y825 A0A1D1VAW0 A0A165JYY0 A0A2C9JS97 D6WZP4 A0A0P6ENL0 A0A0B1TK92 A0A0P6DRR7 A0A0P5VJ77 A0A0U2N329 A0A0P6DF79 A0A0P5F559 K7J3E2 A0A0P5VD00 W6NC52 A0A0P5AQJ9 A0A0P5BJ32 A0A0P5NZL2 A0A0P5N2X2 A0A0P5ATU2 A0A0P5XAL6 Q22633 A0A0N8A5M5 A0A0P5BLJ0 A0A0P5MUW4 A0A0P5Q3X8 A0A0P4ZGJ1 A0A0P5QA72 A0A3B5A4L3 A0A151WHV6 W8B5L7 W8AU74 T1FNN4 R7V6N7 A0A3Q1J7S4 E2AE27 A0A3P8Z521 A0A3S3NZJ3 A0A3B3QJX6 A0A1I7TMB3 A0A0P5GQT2 A0A2G9TNT5 A0A2P8XHR9 A0A0P5SQ41 A0A2P4VGT7 A0A261BV92 E3LZD7 A0A3P8Z5R8 A0A0P6B140 A0A395GUJ1 V4A0B7 A0A3N0YSI0 A0A3P8Z6J3 A0A2G9UN36 Q6AZV8 F1Q986 A0A1B6M4D0 A0A060WF31 A0A0L7QZ05 A0A1S3PI18
EC Number
1.13.11.27
Pubmed
EMBL
NWSH01005678
PCG64062.1
NWSH01007556
PCG62913.1
KQ459580
KPI99320.1
+ More
KQ461196 KPJ06289.1 JTDY01004486 KOB68089.1 AGBW02011783 OWR46152.1 AGBW02011602 OWR46441.1 KPJ06290.1 NWSH01007858 PCG62781.1 AK401453 BAM18075.1 KPI99321.1 NWSH01008285 PCG62625.1 KQ981490 KYN41356.1 QOIP01000009 RLU18348.1 BABH01028340 BABH01028341 KT748795 AMK48576.1 GL888103 EGI67385.1 GL448543 EFN84336.1 NEDP02005541 OWF39134.1 KQ978287 KYM95641.1 GL766526 EFZ15088.1 ODYU01007234 SOQ49835.1 GDHF01033929 JAI18385.1 KK107119 EZA58527.1 ADTU01013709 KZ288204 PBC33226.1 GBYB01014597 JAG84364.1 KQ976736 KYM76052.1 KQ435840 KOX71401.1 UZAF01023597 VDO90544.1 NIVC01002946 PAA54239.1 NIVC01000022 PAA93731.1 NNAY01000645 OXU27215.1 JI172563 ADY45819.1 PDNA01000065 PGH17385.1 BDGG01000005 GAU98781.1 KV407454 KZF26804.1 KQ971372 EFA10456.1 GDIQ01060920 JAN33817.1 KN549647 KHJ96521.1 GDIQ01077658 JAN17079.1 GDIP01099251 JAM04464.1 KR181950 ALJ76674.1 GDIQ01079712 JAN15025.1 GDIQ01267568 JAJ84156.1 GDIP01116891 JAL86823.1 CAVP010058440 CDL94656.1 GDIP01200342 JAJ23060.1 GDIP01198880 JAJ24522.1 GDIQ01142393 JAL09333.1 GDIQ01159286 JAK92439.1 GDIP01198879 JAJ24523.1 GDIP01076462 GDIQ01068717 LRGB01001361 JAM27253.1 JAN26020.1 KZS12608.1 Z50016 GDIP01180137 JAJ43265.1 GDIP01183491 JAJ39911.1 GDIQ01162285 JAK89440.1 GDIQ01140597 JAL11129.1 GDIP01217134 JAJ06268.1 GDIQ01123049 JAL28677.1 KQ983106 KYQ47412.1 GAMC01014147 JAB92408.1 GAMC01014145 JAB92410.1 AMQM01007398 KB097605 ESN93556.1 AMQN01004913 KB294684 ELU14199.1 GL438827 EFN68260.1 NCKU01005195 RWS04895.1 GDIQ01239002 JAK12723.1 KZ357420 PIO59597.1 PYGN01002085 PSN31547.1 GDIQ01092552 JAL59174.1 LFJK02000579 POM36507.1 NIPN01000070 OZG14229.1 DS268419 EFO86670.1 GDIP01021619 JAM82096.1 KZ824460 RAK97773.1 KB201305 ESO97258.1 RJVU01027559 ROL49134.1 KZ345871 PIO71709.1 BC077167 BC165880 AAH77167.1 AAI65880.1 CABZ01049825 CABZ01049826 GEBQ01009219 JAT30758.1 FR904513 CDQ65631.1 KQ414685 KOC63806.1
KQ461196 KPJ06289.1 JTDY01004486 KOB68089.1 AGBW02011783 OWR46152.1 AGBW02011602 OWR46441.1 KPJ06290.1 NWSH01007858 PCG62781.1 AK401453 BAM18075.1 KPI99321.1 NWSH01008285 PCG62625.1 KQ981490 KYN41356.1 QOIP01000009 RLU18348.1 BABH01028340 BABH01028341 KT748795 AMK48576.1 GL888103 EGI67385.1 GL448543 EFN84336.1 NEDP02005541 OWF39134.1 KQ978287 KYM95641.1 GL766526 EFZ15088.1 ODYU01007234 SOQ49835.1 GDHF01033929 JAI18385.1 KK107119 EZA58527.1 ADTU01013709 KZ288204 PBC33226.1 GBYB01014597 JAG84364.1 KQ976736 KYM76052.1 KQ435840 KOX71401.1 UZAF01023597 VDO90544.1 NIVC01002946 PAA54239.1 NIVC01000022 PAA93731.1 NNAY01000645 OXU27215.1 JI172563 ADY45819.1 PDNA01000065 PGH17385.1 BDGG01000005 GAU98781.1 KV407454 KZF26804.1 KQ971372 EFA10456.1 GDIQ01060920 JAN33817.1 KN549647 KHJ96521.1 GDIQ01077658 JAN17079.1 GDIP01099251 JAM04464.1 KR181950 ALJ76674.1 GDIQ01079712 JAN15025.1 GDIQ01267568 JAJ84156.1 GDIP01116891 JAL86823.1 CAVP010058440 CDL94656.1 GDIP01200342 JAJ23060.1 GDIP01198880 JAJ24522.1 GDIQ01142393 JAL09333.1 GDIQ01159286 JAK92439.1 GDIP01198879 JAJ24523.1 GDIP01076462 GDIQ01068717 LRGB01001361 JAM27253.1 JAN26020.1 KZS12608.1 Z50016 GDIP01180137 JAJ43265.1 GDIP01183491 JAJ39911.1 GDIQ01162285 JAK89440.1 GDIQ01140597 JAL11129.1 GDIP01217134 JAJ06268.1 GDIQ01123049 JAL28677.1 KQ983106 KYQ47412.1 GAMC01014147 JAB92408.1 GAMC01014145 JAB92410.1 AMQM01007398 KB097605 ESN93556.1 AMQN01004913 KB294684 ELU14199.1 GL438827 EFN68260.1 NCKU01005195 RWS04895.1 GDIQ01239002 JAK12723.1 KZ357420 PIO59597.1 PYGN01002085 PSN31547.1 GDIQ01092552 JAL59174.1 LFJK02000579 POM36507.1 NIPN01000070 OZG14229.1 DS268419 EFO86670.1 GDIP01021619 JAM82096.1 KZ824460 RAK97773.1 KB201305 ESO97258.1 RJVU01027559 ROL49134.1 KZ345871 PIO71709.1 BC077167 BC165880 AAH77167.1 AAI65880.1 CABZ01049825 CABZ01049826 GEBQ01009219 JAT30758.1 FR904513 CDQ65631.1 KQ414685 KOC63806.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000037510
UP000007151
UP000078541
+ More
UP000279307 UP000005204 UP000095280 UP000007755 UP000008237 UP000242188 UP000078542 UP000085678 UP000053097 UP000005205 UP000242457 UP000078540 UP000005203 UP000053105 UP000038042 UP000268014 UP000215902 UP000192223 UP000215335 UP000005237 UP000224634 UP000186922 UP000076632 UP000076420 UP000007266 UP000002358 UP000025227 UP000076858 UP000001940 UP000261400 UP000075809 UP000015101 UP000014760 UP000265040 UP000000311 UP000265140 UP000285301 UP000261540 UP000095282 UP000245037 UP000237256 UP000216463 UP000008281 UP000249402 UP000030746 UP000000437 UP000193380 UP000053825 UP000087266
UP000279307 UP000005204 UP000095280 UP000007755 UP000008237 UP000242188 UP000078542 UP000085678 UP000053097 UP000005205 UP000242457 UP000078540 UP000005203 UP000053105 UP000038042 UP000268014 UP000215902 UP000192223 UP000215335 UP000005237 UP000224634 UP000186922 UP000076632 UP000076420 UP000007266 UP000002358 UP000025227 UP000076858 UP000001940 UP000261400 UP000075809 UP000015101 UP000014760 UP000265040 UP000000311 UP000265140 UP000285301 UP000261540 UP000095282 UP000245037 UP000237256 UP000216463 UP000008281 UP000249402 UP000030746 UP000000437 UP000193380 UP000053825 UP000087266
PRIDE
Pfam
PF00903 Glyoxalase
Interpro
SUPFAM
SSF54593
SSF54593
Gene 3D
ProteinModelPortal
A0A2A4IY11
A0A2A4IUV6
A0A194Q7D9
A0A194QLP7
A0A0L7KYG5
A0A212EXC3
+ More
A0A212EYA1 A0A194QMV1 A0A2A4ISS0 I4DJI5 A0A194Q189 A0A2A4IS65 A0A195FM03 A0A3L8DEL4 H9J6X0 A0A140DKW1 A0A1I8IJD7 F4WEM2 E2BIZ1 A0A210PRQ4 A0A195C642 E9IWI6 A0A2H1W9R5 A0A0K8TWD3 A0A1S3JV90 A0A026WR60 A0A158NF97 A0A2A3EPS1 A0A0C9RMG1 A0A195AVG6 A0A088ACK9 A0A0N0U440 A0A0N4XB01 A0A267E053 A0A267H602 A0A1W4WLA9 A0A232F978 A0A1I8IMR3 F1L6R5 A0A2H2IKY3 A0A2B7Y825 A0A1D1VAW0 A0A165JYY0 A0A2C9JS97 D6WZP4 A0A0P6ENL0 A0A0B1TK92 A0A0P6DRR7 A0A0P5VJ77 A0A0U2N329 A0A0P6DF79 A0A0P5F559 K7J3E2 A0A0P5VD00 W6NC52 A0A0P5AQJ9 A0A0P5BJ32 A0A0P5NZL2 A0A0P5N2X2 A0A0P5ATU2 A0A0P5XAL6 Q22633 A0A0N8A5M5 A0A0P5BLJ0 A0A0P5MUW4 A0A0P5Q3X8 A0A0P4ZGJ1 A0A0P5QA72 A0A3B5A4L3 A0A151WHV6 W8B5L7 W8AU74 T1FNN4 R7V6N7 A0A3Q1J7S4 E2AE27 A0A3P8Z521 A0A3S3NZJ3 A0A3B3QJX6 A0A1I7TMB3 A0A0P5GQT2 A0A2G9TNT5 A0A2P8XHR9 A0A0P5SQ41 A0A2P4VGT7 A0A261BV92 E3LZD7 A0A3P8Z5R8 A0A0P6B140 A0A395GUJ1 V4A0B7 A0A3N0YSI0 A0A3P8Z6J3 A0A2G9UN36 Q6AZV8 F1Q986 A0A1B6M4D0 A0A060WF31 A0A0L7QZ05 A0A1S3PI18
A0A212EYA1 A0A194QMV1 A0A2A4ISS0 I4DJI5 A0A194Q189 A0A2A4IS65 A0A195FM03 A0A3L8DEL4 H9J6X0 A0A140DKW1 A0A1I8IJD7 F4WEM2 E2BIZ1 A0A210PRQ4 A0A195C642 E9IWI6 A0A2H1W9R5 A0A0K8TWD3 A0A1S3JV90 A0A026WR60 A0A158NF97 A0A2A3EPS1 A0A0C9RMG1 A0A195AVG6 A0A088ACK9 A0A0N0U440 A0A0N4XB01 A0A267E053 A0A267H602 A0A1W4WLA9 A0A232F978 A0A1I8IMR3 F1L6R5 A0A2H2IKY3 A0A2B7Y825 A0A1D1VAW0 A0A165JYY0 A0A2C9JS97 D6WZP4 A0A0P6ENL0 A0A0B1TK92 A0A0P6DRR7 A0A0P5VJ77 A0A0U2N329 A0A0P6DF79 A0A0P5F559 K7J3E2 A0A0P5VD00 W6NC52 A0A0P5AQJ9 A0A0P5BJ32 A0A0P5NZL2 A0A0P5N2X2 A0A0P5ATU2 A0A0P5XAL6 Q22633 A0A0N8A5M5 A0A0P5BLJ0 A0A0P5MUW4 A0A0P5Q3X8 A0A0P4ZGJ1 A0A0P5QA72 A0A3B5A4L3 A0A151WHV6 W8B5L7 W8AU74 T1FNN4 R7V6N7 A0A3Q1J7S4 E2AE27 A0A3P8Z521 A0A3S3NZJ3 A0A3B3QJX6 A0A1I7TMB3 A0A0P5GQT2 A0A2G9TNT5 A0A2P8XHR9 A0A0P5SQ41 A0A2P4VGT7 A0A261BV92 E3LZD7 A0A3P8Z5R8 A0A0P6B140 A0A395GUJ1 V4A0B7 A0A3N0YSI0 A0A3P8Z6J3 A0A2G9UN36 Q6AZV8 F1Q986 A0A1B6M4D0 A0A060WF31 A0A0L7QZ05 A0A1S3PI18
PDB
1SQI
E-value=4.02559e-15,
Score=191
Ontologies
PATHWAY
GO
PANTHER
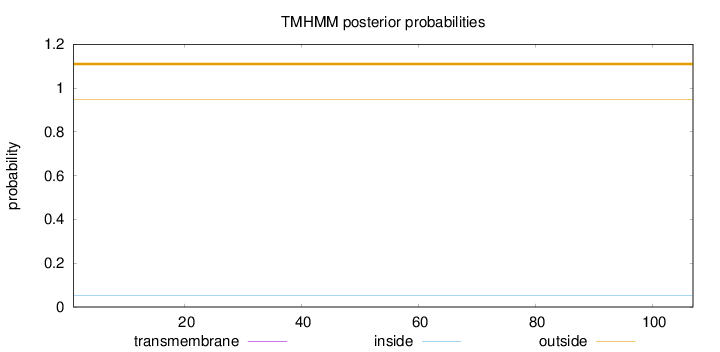
Topology
Length:
107
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00132
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05263
outside
1 - 107
Population Genetic Test Statistics
Pi
137.449887
Theta
71.406709
Tajima's D
1.479351
CLR
0
CSRT
0.780260986950652
Interpretation
Uncertain
