Gene
KWMTBOMO04346
Pre Gene Modal
BGIBMGA005261
Annotation
PREDICTED:_4-hydroxyphenylpyruvate_dioxygenase-like_[Amyelois_transitella]
Full name
4-hydroxyphenylpyruvate dioxygenase
Location in the cell
Cytoplasmic Reliability : 1.652
Sequence
CDS
ATGGCGGACGGCACTCTAGAAGATTCTGTCGCCTGGTACGAAAAGAATTTAAATATGCACCGGTTCTGGTGCGTGGACTACAGCCACGATTTAGTGCCCTATTCCTGCATAAATTCTGCATCAGTAATAAATCAAGATGAAACTATATTGTTGTCTATGAACGAAGCAGCGAAAGGATTGCGGCCTTCGAGCAAAGCGTCAGAGTTCGTTAAAGCGCTCGGTACGTCTGGTGTTGAACACATAGCACTGTACACTGATGACATTGTAAATACTATGCAAAACTTAAAAAAACGCGGTGCCGAAATAATAACGTGGCCGTCAACATACTACGAGATAATCAGAGAAAAACTGACCAAGAGTACAGTGAAAGTCGCAGAGAGCCTAACCGCTCTGCAGGAAAATAATATCCTGATCGACTTCGATGAAAGAGGCTACATGCTGCAGGCCTTCACTAAGCACGTACAAGCCCGACCGACGGTTTTCATAGAAATTTTGCAGAGGAGAAACCATAGGGGATTCGGCGCTATGAATTACAAATGGGTTTTCGAAGCGATCGAGCGATTGGAAACCGCTAAAGATATTAAAAGCGATGAAAGTAAGACAGACAAGTGA
Protein
MADGTLEDSVAWYEKNLNMHRFWCVDYSHDLVPYSCINSASVINQDETILLSMNEAAKGLRPSSKASEFVKALGTSGVEHIALYTDDIVNTMQNLKKRGAEIITWPSTYYEIIREKLTKSTVKVAESLTALQENNILIDFDERGYMLQAFTKHVQARPTVFIEILQRRNHRGFGAMNYKWVFEAIERLETAKDIKSDESKTDK
Summary
Cofactor
Fe cation
Similarity
Belongs to the 4HPPD family.
Feature
chain 4-hydroxyphenylpyruvate dioxygenase
Uniprot
A0A2A4IUV6
A0A2H1W9R5
A0A194QLP7
A0A194Q7D9
A0A212EYA1
H9J6W9
+ More
A0A0L7KYG5 A0A0L7KJD7 A0A0P4Z082 A0A0P6EY08 A0A0P5CXZ8 A0A0P5QA72 A0A0P4ZWI9 A0A0P5AIL6 A0A0S7HZE5 A0A2A4ISS0 A0A0P4YSK2 A0A0P5UKN1 A0A0N8A5M5 A0A0P5AQJ9 A0A0P5BLJ0 A0A0P5T823 A0A0P5S574 A0A0P5SLR8 A0A0P5BJ32 M4ALC3 A0A147A5U7 A0A0R1E8T5 A0A2A4IS65 A0A087YG21 A0A0N8A506 A0A0N8BPL6 A0A0P5Q3X8 A0A0P5NZL2 A0A0P5XAL6 A0A0P5F559 A0A0P5WLA1 A0A0P5M1B1 A0A0P5HFL7 A0A0P5SQ41 A0A1L8E2X1 A0A3P9P138 A0A132AM89 A0A0P5GQT2 A0A0H5RAY7 A0A0P5RFX9 A0A0P5HC09 A0A1W4Y6X9 A0A1Y3EXQ0 A0A0P6GU61 A0A091SPB8 A0A1I8D1E5 A0A1W4YG50 E9H9X2 A0A0P5MDG0 A0A2P2HW87 Q961W1 A0A182SWY5 A0A0P6B140 I3JXN2 A0A3Q2DQN4 A0A093J154 A0A182UNA3 B3M9A0 A0A0V1I7J2 A0A182UEF0 A0A3Q2VR45 A0A2Y9D3F6 A0A182LI36 Q7Q2T3 A0A3P8PMU8 M7C925 A0A3P9BSL5 A0A3P9BSH0 A0A336MHT6 G3PDN8 A0A3Q2DRI1 A0A0P4ZGJ1 A0A0P4W1F5 A0A1B2G378 A0A182JAR9 A0A0N4ZSG9 A0A0P6DF79 B3NIM3 B4IUG7 B4N6M5 A0A3Q0T349 A0A0V1G5N9 A0A0V1N298 D6WZP4 A0A0C9RYZ0 A0A2M4AGP9 A0A182MNP5 G3PDP2 A0A0J9UQ37 Q9VPF3 W5JMN3 A0A218UNA2 A0A0U2N329 T1E8I4
A0A0L7KYG5 A0A0L7KJD7 A0A0P4Z082 A0A0P6EY08 A0A0P5CXZ8 A0A0P5QA72 A0A0P4ZWI9 A0A0P5AIL6 A0A0S7HZE5 A0A2A4ISS0 A0A0P4YSK2 A0A0P5UKN1 A0A0N8A5M5 A0A0P5AQJ9 A0A0P5BLJ0 A0A0P5T823 A0A0P5S574 A0A0P5SLR8 A0A0P5BJ32 M4ALC3 A0A147A5U7 A0A0R1E8T5 A0A2A4IS65 A0A087YG21 A0A0N8A506 A0A0N8BPL6 A0A0P5Q3X8 A0A0P5NZL2 A0A0P5XAL6 A0A0P5F559 A0A0P5WLA1 A0A0P5M1B1 A0A0P5HFL7 A0A0P5SQ41 A0A1L8E2X1 A0A3P9P138 A0A132AM89 A0A0P5GQT2 A0A0H5RAY7 A0A0P5RFX9 A0A0P5HC09 A0A1W4Y6X9 A0A1Y3EXQ0 A0A0P6GU61 A0A091SPB8 A0A1I8D1E5 A0A1W4YG50 E9H9X2 A0A0P5MDG0 A0A2P2HW87 Q961W1 A0A182SWY5 A0A0P6B140 I3JXN2 A0A3Q2DQN4 A0A093J154 A0A182UNA3 B3M9A0 A0A0V1I7J2 A0A182UEF0 A0A3Q2VR45 A0A2Y9D3F6 A0A182LI36 Q7Q2T3 A0A3P8PMU8 M7C925 A0A3P9BSL5 A0A3P9BSH0 A0A336MHT6 G3PDN8 A0A3Q2DRI1 A0A0P4ZGJ1 A0A0P4W1F5 A0A1B2G378 A0A182JAR9 A0A0N4ZSG9 A0A0P6DF79 B3NIM3 B4IUG7 B4N6M5 A0A3Q0T349 A0A0V1G5N9 A0A0V1N298 D6WZP4 A0A0C9RYZ0 A0A2M4AGP9 A0A182MNP5 G3PDP2 A0A0J9UQ37 Q9VPF3 W5JMN3 A0A218UNA2 A0A0U2N329 T1E8I4
Pubmed
26354079
22118469
19121390
26227816
23542700
17994087
+ More
17550304 26555130 26829753 21292972 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25186727 20966253 12364791 14747013 17210077 23624526 27129103 27476595 18362917 19820115 22936249 20920257 23761445
17550304 26555130 26829753 21292972 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25186727 20966253 12364791 14747013 17210077 23624526 27129103 27476595 18362917 19820115 22936249 20920257 23761445
EMBL
NWSH01007556
PCG62913.1
ODYU01007234
SOQ49835.1
KQ461196
KPJ06289.1
+ More
KQ459580 KPI99320.1 AGBW02011602 OWR46441.1 BABH01028342 JTDY01004486 KOB68089.1 JTDY01009626 KOB63210.1 GDIP01224287 JAI99114.1 GDIQ01056025 JAN38712.1 GDIP01180138 JAJ43264.1 GDIQ01123049 JAL28677.1 GDIP01208738 JAJ14664.1 GDIP01211946 JAJ11456.1 GBYX01436353 JAO44985.1 NWSH01007858 PCG62781.1 GDIP01224286 JAI99115.1 GDIP01111603 JAL92111.1 GDIP01180137 JAJ43265.1 GDIP01200342 JAJ23060.1 GDIP01183491 JAJ39911.1 GDIP01131345 JAL72369.1 GDIQ01096560 JAL55166.1 GDIP01138193 JAL65521.1 GDIP01198880 JAJ24522.1 GCES01012400 JAR73923.1 CH896779 KRK05659.1 NWSH01008285 PCG62625.1 AYCK01000982 GDIP01181889 JAJ41513.1 GDIQ01157331 JAK94394.1 GDIQ01140597 JAL11129.1 GDIQ01142393 JAL09333.1 GDIP01076462 GDIQ01068717 LRGB01001361 JAM27253.1 JAN26020.1 KZS12608.1 GDIQ01267568 JAJ84156.1 GDIQ01247403 GDIP01084650 JAK04322.1 JAM19065.1 GDIQ01162286 JAK89439.1 GDIQ01250658 JAK01067.1 GDIQ01092552 JAL59174.1 GFDF01001239 JAV12845.1 JXLN01018432 KPM11969.1 GDIQ01239002 JAK12723.1 HACM01010903 CRZ11345.1 GDIQ01106498 JAL45228.1 GDIQ01228910 JAK22815.1 LVZM01000231 OUC49891.1 GDIQ01028672 JAN66065.1 KK478773 KFQ59855.1 GL732609 EFX71536.1 GDIQ01157330 JAK94395.1 IACF01000265 LAB66059.1 AE014296 AY047523 AAF51602.1 AAK77255.1 GDIP01021619 JAM82096.1 AERX01013659 KK566660 KFW04739.1 CH902618 EDV40084.2 JYDP01000003 KRZ18404.1 AAAB01008968 EAA13250.3 KB523138 EMP37107.1 UFQT01001230 SSX29585.1 GDIP01217134 JAJ06268.1 GDKW01001094 JAI55501.1 KX572142 ANZ03346.1 GDIQ01079712 JAN15025.1 CH954178 EDV52519.1 CH891904 EDX00031.1 CH964161 EDW80014.2 JYDT01000002 JYDV01000010 KRY93426.1 KRZ43398.1 JYDO01000014 KRZ78141.1 KQ971372 EFA10456.1 GBYB01014595 JAG84362.1 GGFK01006635 MBW39956.1 AXCM01006965 CM002912 KMZ00796.1 BT012317 AAF51601.1 AAS77442.1 ADMH02001127 ETN64019.1 MUZQ01000225 OWK54910.1 KR181950 ALJ76674.1 GAMD01002738 JAA98852.1
KQ459580 KPI99320.1 AGBW02011602 OWR46441.1 BABH01028342 JTDY01004486 KOB68089.1 JTDY01009626 KOB63210.1 GDIP01224287 JAI99114.1 GDIQ01056025 JAN38712.1 GDIP01180138 JAJ43264.1 GDIQ01123049 JAL28677.1 GDIP01208738 JAJ14664.1 GDIP01211946 JAJ11456.1 GBYX01436353 JAO44985.1 NWSH01007858 PCG62781.1 GDIP01224286 JAI99115.1 GDIP01111603 JAL92111.1 GDIP01180137 JAJ43265.1 GDIP01200342 JAJ23060.1 GDIP01183491 JAJ39911.1 GDIP01131345 JAL72369.1 GDIQ01096560 JAL55166.1 GDIP01138193 JAL65521.1 GDIP01198880 JAJ24522.1 GCES01012400 JAR73923.1 CH896779 KRK05659.1 NWSH01008285 PCG62625.1 AYCK01000982 GDIP01181889 JAJ41513.1 GDIQ01157331 JAK94394.1 GDIQ01140597 JAL11129.1 GDIQ01142393 JAL09333.1 GDIP01076462 GDIQ01068717 LRGB01001361 JAM27253.1 JAN26020.1 KZS12608.1 GDIQ01267568 JAJ84156.1 GDIQ01247403 GDIP01084650 JAK04322.1 JAM19065.1 GDIQ01162286 JAK89439.1 GDIQ01250658 JAK01067.1 GDIQ01092552 JAL59174.1 GFDF01001239 JAV12845.1 JXLN01018432 KPM11969.1 GDIQ01239002 JAK12723.1 HACM01010903 CRZ11345.1 GDIQ01106498 JAL45228.1 GDIQ01228910 JAK22815.1 LVZM01000231 OUC49891.1 GDIQ01028672 JAN66065.1 KK478773 KFQ59855.1 GL732609 EFX71536.1 GDIQ01157330 JAK94395.1 IACF01000265 LAB66059.1 AE014296 AY047523 AAF51602.1 AAK77255.1 GDIP01021619 JAM82096.1 AERX01013659 KK566660 KFW04739.1 CH902618 EDV40084.2 JYDP01000003 KRZ18404.1 AAAB01008968 EAA13250.3 KB523138 EMP37107.1 UFQT01001230 SSX29585.1 GDIP01217134 JAJ06268.1 GDKW01001094 JAI55501.1 KX572142 ANZ03346.1 GDIQ01079712 JAN15025.1 CH954178 EDV52519.1 CH891904 EDX00031.1 CH964161 EDW80014.2 JYDT01000002 JYDV01000010 KRY93426.1 KRZ43398.1 JYDO01000014 KRZ78141.1 KQ971372 EFA10456.1 GBYB01014595 JAG84362.1 GGFK01006635 MBW39956.1 AXCM01006965 CM002912 KMZ00796.1 BT012317 AAF51601.1 AAS77442.1 ADMH02001127 ETN64019.1 MUZQ01000225 OWK54910.1 KR181950 ALJ76674.1 GAMD01002738 JAA98852.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000005204
UP000037510
+ More
UP000002852 UP000265000 UP000002282 UP000028760 UP000076858 UP000242638 UP000192224 UP000243006 UP000095286 UP000000305 UP000000803 UP000075901 UP000005207 UP000265020 UP000075903 UP000007801 UP000055024 UP000075902 UP000264840 UP000076407 UP000075882 UP000007062 UP000265100 UP000031443 UP000265160 UP000007635 UP000075880 UP000038045 UP000008711 UP000007798 UP000261340 UP000054826 UP000054995 UP000054843 UP000007266 UP000075883 UP000000673 UP000197619
UP000002852 UP000265000 UP000002282 UP000028760 UP000076858 UP000242638 UP000192224 UP000243006 UP000095286 UP000000305 UP000000803 UP000075901 UP000005207 UP000265020 UP000075903 UP000007801 UP000055024 UP000075902 UP000264840 UP000076407 UP000075882 UP000007062 UP000265100 UP000031443 UP000265160 UP000007635 UP000075880 UP000038045 UP000008711 UP000007798 UP000261340 UP000054826 UP000054995 UP000054843 UP000007266 UP000075883 UP000000673 UP000197619
Pfam
PF00903 Glyoxalase
Interpro
SUPFAM
SSF54593
SSF54593
Gene 3D
ProteinModelPortal
A0A2A4IUV6
A0A2H1W9R5
A0A194QLP7
A0A194Q7D9
A0A212EYA1
H9J6W9
+ More
A0A0L7KYG5 A0A0L7KJD7 A0A0P4Z082 A0A0P6EY08 A0A0P5CXZ8 A0A0P5QA72 A0A0P4ZWI9 A0A0P5AIL6 A0A0S7HZE5 A0A2A4ISS0 A0A0P4YSK2 A0A0P5UKN1 A0A0N8A5M5 A0A0P5AQJ9 A0A0P5BLJ0 A0A0P5T823 A0A0P5S574 A0A0P5SLR8 A0A0P5BJ32 M4ALC3 A0A147A5U7 A0A0R1E8T5 A0A2A4IS65 A0A087YG21 A0A0N8A506 A0A0N8BPL6 A0A0P5Q3X8 A0A0P5NZL2 A0A0P5XAL6 A0A0P5F559 A0A0P5WLA1 A0A0P5M1B1 A0A0P5HFL7 A0A0P5SQ41 A0A1L8E2X1 A0A3P9P138 A0A132AM89 A0A0P5GQT2 A0A0H5RAY7 A0A0P5RFX9 A0A0P5HC09 A0A1W4Y6X9 A0A1Y3EXQ0 A0A0P6GU61 A0A091SPB8 A0A1I8D1E5 A0A1W4YG50 E9H9X2 A0A0P5MDG0 A0A2P2HW87 Q961W1 A0A182SWY5 A0A0P6B140 I3JXN2 A0A3Q2DQN4 A0A093J154 A0A182UNA3 B3M9A0 A0A0V1I7J2 A0A182UEF0 A0A3Q2VR45 A0A2Y9D3F6 A0A182LI36 Q7Q2T3 A0A3P8PMU8 M7C925 A0A3P9BSL5 A0A3P9BSH0 A0A336MHT6 G3PDN8 A0A3Q2DRI1 A0A0P4ZGJ1 A0A0P4W1F5 A0A1B2G378 A0A182JAR9 A0A0N4ZSG9 A0A0P6DF79 B3NIM3 B4IUG7 B4N6M5 A0A3Q0T349 A0A0V1G5N9 A0A0V1N298 D6WZP4 A0A0C9RYZ0 A0A2M4AGP9 A0A182MNP5 G3PDP2 A0A0J9UQ37 Q9VPF3 W5JMN3 A0A218UNA2 A0A0U2N329 T1E8I4
A0A0L7KYG5 A0A0L7KJD7 A0A0P4Z082 A0A0P6EY08 A0A0P5CXZ8 A0A0P5QA72 A0A0P4ZWI9 A0A0P5AIL6 A0A0S7HZE5 A0A2A4ISS0 A0A0P4YSK2 A0A0P5UKN1 A0A0N8A5M5 A0A0P5AQJ9 A0A0P5BLJ0 A0A0P5T823 A0A0P5S574 A0A0P5SLR8 A0A0P5BJ32 M4ALC3 A0A147A5U7 A0A0R1E8T5 A0A2A4IS65 A0A087YG21 A0A0N8A506 A0A0N8BPL6 A0A0P5Q3X8 A0A0P5NZL2 A0A0P5XAL6 A0A0P5F559 A0A0P5WLA1 A0A0P5M1B1 A0A0P5HFL7 A0A0P5SQ41 A0A1L8E2X1 A0A3P9P138 A0A132AM89 A0A0P5GQT2 A0A0H5RAY7 A0A0P5RFX9 A0A0P5HC09 A0A1W4Y6X9 A0A1Y3EXQ0 A0A0P6GU61 A0A091SPB8 A0A1I8D1E5 A0A1W4YG50 E9H9X2 A0A0P5MDG0 A0A2P2HW87 Q961W1 A0A182SWY5 A0A0P6B140 I3JXN2 A0A3Q2DQN4 A0A093J154 A0A182UNA3 B3M9A0 A0A0V1I7J2 A0A182UEF0 A0A3Q2VR45 A0A2Y9D3F6 A0A182LI36 Q7Q2T3 A0A3P8PMU8 M7C925 A0A3P9BSL5 A0A3P9BSH0 A0A336MHT6 G3PDN8 A0A3Q2DRI1 A0A0P4ZGJ1 A0A0P4W1F5 A0A1B2G378 A0A182JAR9 A0A0N4ZSG9 A0A0P6DF79 B3NIM3 B4IUG7 B4N6M5 A0A3Q0T349 A0A0V1G5N9 A0A0V1N298 D6WZP4 A0A0C9RYZ0 A0A2M4AGP9 A0A182MNP5 G3PDP2 A0A0J9UQ37 Q9VPF3 W5JMN3 A0A218UNA2 A0A0U2N329 T1E8I4
PDB
1SQI
E-value=9.74689e-46,
Score=459
Ontologies
PANTHER
Topology
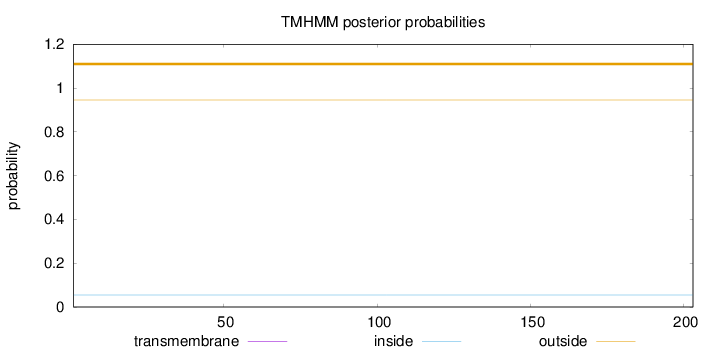
Length:
203
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00235
Exp number, first 60 AAs:
0.00178
Total prob of N-in:
0.05460
outside
1 - 203
Population Genetic Test Statistics
Pi
176.20596
Theta
18.030943
Tajima's D
0.625377
CLR
0.332588
CSRT
0.554022298885056
Interpretation
Uncertain
