Gene
KWMTBOMO04343
Pre Gene Modal
BGIBMGA005263
Annotation
PREDICTED:_probable_serine_hydrolase_[Amyelois_transitella]
Full name
Probable serine hydrolase
Alternative Name
Kraken protein
Location in the cell
Cytoplasmic Reliability : 4.056
Sequence
CDS
ATGGAAGAGAAAACGAATGGATATCAAAACCATGAAGAACTCCCGCCTGGCTACACAGTCGAGGAAGTCGAAATACCAGTGCCCTGGGGTCACGTCGCCGGCCGTTGGTGGGGACCGCGGAACATACAGCCCATGATCGCGATCCACGGCTGGCAGGACAACGCCGGAACTTGGGACAACCTCATTCCCTTGTTACCGGTTAACACCTCGGTCCTCTGCATAGACTTGCCGGGTCACGGCCACTCCTCTCACTACCCCAAAGGTATGATTTACTACGTTTTCTGGGACGGAATCGTTTTGATCCGACGAATCGTGAAGCACTTCAAATGGGAGAAGGTACAGTTGATGGGACATTCCCTGGGCGGCGCCCTGAGCTTCATGTACGCGGCCTCGTTTCCCGACGATGTCGATAAGATTATATGCATCGATATCGCGAGTCCGGCTGTAAGGGAACCGTCTAATATGGTCAAAACAGCCGGATGGGGTATGGACAAGCTGCTCGAGTATGAAACATTGAAGGAAGATAAGATACCGTGTTACGAATACGAGGAAATGATAGATATAGTATGCGATGCGTACAAAGGATCCGTTTCGAAGCGTAACTGCCGCGTGCTGATGAGACGGGGCATGATGCCGACCCCGGCGCATATGAACAAGAAGGGTTACTTCTTCAAAAGGGATCCGCGCTTGAAAGTGGCAGGGTTGGCGATGATGTCGATCGAGACCGCTTTAGAGTTCGCCGGCAAGGTCAAGTGCAAAGTGCTGAATTTGAGGGCGTTGCCCGGCCAGGGCTGGGAGAGGCTAGATTATTACATGAAGGTAGTTGAAAAATTGAGAGAAACCACCGATGTCCAGTACGTCGAGGTGCAGGGAACGCATCACGTTCAATTAGAATCGCCCGAAAACATCGTTGAACTCATAGAAGAGTTCTTAGAGCTTTATTAA
Protein
MEEKTNGYQNHEELPPGYTVEEVEIPVPWGHVAGRWWGPRNIQPMIAIHGWQDNAGTWDNLIPLLPVNTSVLCIDLPGHGHSSHYPKGMIYYVFWDGIVLIRRIVKHFKWEKVQLMGHSLGGALSFMYAASFPDDVDKIICIDIASPAVREPSNMVKTAGWGMDKLLEYETLKEDKIPCYEYEEMIDIVCDAYKGSVSKRNCRVLMRRGMMPTPAHMNKKGYFFKRDPRLKVAGLAMMSIETALEFAGKVKCKVLNLRALPGQGWERLDYYMKVVEKLRETTDVQYVEVQGTHHVQLESPENIVELIEEFLELY
Summary
Similarity
Belongs to the intermediate filament family.
Belongs to the AB hydrolase superfamily.
Belongs to the AB hydrolase superfamily.
Keywords
Complete proteome
Detoxification
Developmental protein
Digestion
Hydrolase
Reference proteome
Serine esterase
Feature
chain Probable serine hydrolase
Uniprot
H9J6X1
A0A194QL80
A0A194Q1W8
A0A2H1W9W4
A0A2A4JQD2
A0A212EY73
+ More
A0A2A4ITD1 A0A0L7L2Q8 A0A3S2M599 B0VZX9 B0X1Q9 A0A1Q3FAX7 A0A1Q3FE02 A0A1S4H3F5 A0A182X7A8 A0A2M3ZCK3 A0A182I0T4 A0A182Q612 A0A034VZL3 A0A182VSV4 A0A0K8W4M4 A0A0A1WMK6 E0VGA1 A0A2M4APK9 A0A2M4APK7 Q7PME1 A0A182YHF7 W5J9D0 A0A2M4BVJ2 E2BMU7 W8BTN0 A0A182RAC0 A0A1J1I4D1 Q16K76 Q16IQ5 A0A336MMY0 A0A336MEG2 A0A182H8W0 A0A1B0A5U1 A0A1I8Q8J6 A0A1L8EIA2 A0A1L8EIG7 A0A1B0AS70 D3TNU0 A0A1A9VH29 A0A1A9X0Q8 A0A0M4DZD8 B4KF70 A0A0Q9WIQ8 A0A336N3V9 B4LTY2 A0A0L0CD77 A0A3B0JZK8 A0A1W4V2Q3 A0A1A9Y0D2 B3MM66 K7JF39 A0A182KJV4 A0A0P8XUU7 B4JDU1 Q29M81 B0FE52 A0A3B0JUG6 D6WZ04 T1PN51 T1PAW0 A0A182M175 A0A1W4WGQ0 B4P2Y4 B0FE49 B2ZI86 B4N0Q1 B2ZI84 B3N895 A0A1B6EEB3 V9IEZ5 B2ZIC5 M9PBM0 O18391 A0A2M4BW91 A0A182H5N0 A0A195BE09 A0A1B6JMD8 A0A2P8YJF8 A0A1Y1LDA6 A0A1Y1LD67 A0A2H8TEJ1 A0A1Y1LDA2 E9J8P2 E2AA78 A0A182N8T6 A0A3L8DA60 A0A182SKB6 A0A0A9ZHH3 J9K888 A0A146LCD4 A0A3L8DB58 A0A067QTY7 A0A0J7KZJ8 A0A182KA99
A0A2A4ITD1 A0A0L7L2Q8 A0A3S2M599 B0VZX9 B0X1Q9 A0A1Q3FAX7 A0A1Q3FE02 A0A1S4H3F5 A0A182X7A8 A0A2M3ZCK3 A0A182I0T4 A0A182Q612 A0A034VZL3 A0A182VSV4 A0A0K8W4M4 A0A0A1WMK6 E0VGA1 A0A2M4APK9 A0A2M4APK7 Q7PME1 A0A182YHF7 W5J9D0 A0A2M4BVJ2 E2BMU7 W8BTN0 A0A182RAC0 A0A1J1I4D1 Q16K76 Q16IQ5 A0A336MMY0 A0A336MEG2 A0A182H8W0 A0A1B0A5U1 A0A1I8Q8J6 A0A1L8EIA2 A0A1L8EIG7 A0A1B0AS70 D3TNU0 A0A1A9VH29 A0A1A9X0Q8 A0A0M4DZD8 B4KF70 A0A0Q9WIQ8 A0A336N3V9 B4LTY2 A0A0L0CD77 A0A3B0JZK8 A0A1W4V2Q3 A0A1A9Y0D2 B3MM66 K7JF39 A0A182KJV4 A0A0P8XUU7 B4JDU1 Q29M81 B0FE52 A0A3B0JUG6 D6WZ04 T1PN51 T1PAW0 A0A182M175 A0A1W4WGQ0 B4P2Y4 B0FE49 B2ZI86 B4N0Q1 B2ZI84 B3N895 A0A1B6EEB3 V9IEZ5 B2ZIC5 M9PBM0 O18391 A0A2M4BW91 A0A182H5N0 A0A195BE09 A0A1B6JMD8 A0A2P8YJF8 A0A1Y1LDA6 A0A1Y1LD67 A0A2H8TEJ1 A0A1Y1LDA2 E9J8P2 E2AA78 A0A182N8T6 A0A3L8DA60 A0A182SKB6 A0A0A9ZHH3 J9K888 A0A146LCD4 A0A3L8DB58 A0A067QTY7 A0A0J7KZJ8 A0A182KA99
EC Number
3.1.-.-
Pubmed
19121390
26354079
22118469
26227816
12364791
25348373
+ More
25830018 20566863 25244985 20920257 23761445 20798317 24495485 17510324 26483478 20353571 17994087 26108605 20075255 20966253 15632085 18288436 18362917 19820115 25315136 17550304 18560591 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 9831651 12537569 29403074 28004739 21282665 30249741 25401762 26823975 24845553
25830018 20566863 25244985 20920257 23761445 20798317 24495485 17510324 26483478 20353571 17994087 26108605 20075255 20966253 15632085 18288436 18362917 19820115 25315136 17550304 18560591 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 9831651 12537569 29403074 28004739 21282665 30249741 25401762 26823975 24845553
EMBL
BABH01028335
KQ461196
KPJ06292.1
KQ459580
KPI99323.1
ODYU01007234
+ More
SOQ49837.1 NWSH01000895 PCG73653.1 AGBW02011602 OWR46443.1 NWSH01008552 PCG62504.1 JTDY01003429 KOB69569.1 RSAL01000030 RVE51714.1 DS231816 EDS37551.1 DS232267 EDS38787.1 GFDL01010319 JAV24726.1 GFDL01009252 JAV25793.1 AAAB01008980 GGFM01005481 MBW26232.1 APCN01005264 AXCN02001866 GAKP01011642 JAC47310.1 GDHF01006504 JAI45810.1 GBXI01014028 JAD00264.1 DS235134 EEB12407.1 GGFK01009389 MBW42710.1 GGFK01009404 MBW42725.1 EAA13959.5 ADMH02002079 ETN59455.1 GGFJ01007627 MBW56768.1 GL449347 EFN82982.1 GAMC01004018 JAC02538.1 CVRI01000037 CRK93241.1 CH477974 EAT34690.1 CH478060 EAT34149.1 UFQS01001355 UFQT01001355 SSX10497.1 SSX30183.1 UFQT01000874 SSX27761.1 JXUM01119439 JXUM01119440 JXUM01119441 JXUM01119442 JXUM01119443 KQ566290 KXJ70258.1 GFDG01000415 JAV18384.1 GFDG01000416 JAV18383.1 JXJN01002686 EZ423092 ADD19368.1 CP012523 ALC37928.1 CH933807 EDW13053.2 CH940649 KRF81991.1 UFQS01004235 UFQT01004235 SSX16266.1 SSX35587.1 EDW65035.2 JRES01000668 KNC29414.1 OUUW01000004 SPP79119.1 CH902620 EDV30881.1 AAZX01003293 KPU73097.1 CH916368 EDW03461.1 CH379060 EAL33813.1 EU310340 ABY55448.1 SPP79120.1 KQ971363 EFA07837.2 KA649383 AFP64012.1 KA645901 AFP60530.1 AXCM01000224 CM000157 EDW87191.1 EU310337 EU310338 EU310339 EU310341 ABY55445.1 ABY55446.1 ABY55447.1 ABY55449.1 EU670477 ACD56183.1 CH963920 EDW77664.2 EU670475 EU670476 EU670478 CM000361 CM002910 ACD56181.1 ACD56182.1 ACD56184.1 EDX03271.1 KMY87383.1 CH954177 EDV57282.2 GEDC01012856 GEDC01001009 JAS24442.1 JAS36289.1 JR042514 AEY59630.1 EU670516 CH480829 ACD56185.1 EDW45450.1 AE014134 AGB92384.1 AJ000516 AY095092 AY118303 GGFJ01008133 MBW57274.1 JXUM01112104 KQ565562 KXJ70883.1 KQ976511 KYM82449.1 GECU01019787 GECU01010408 GECU01007454 GECU01004744 JAS87919.1 JAS97298.1 JAT00253.1 JAT02963.1 PYGN01000551 PSN44389.1 GEZM01058957 JAV71603.1 GEZM01058958 JAV71602.1 GFXV01000665 MBW12470.1 GEZM01058956 JAV71604.1 GL769036 EFZ10818.1 GL438030 EFN69658.1 QOIP01000010 RLU17385.1 GBHO01000341 GBRD01000894 JAG43263.1 JAG64927.1 ABLF02021049 GDHC01012626 JAQ06003.1 RLU17382.1 KK853018 KDR12428.1 LBMM01001627 KMQ95982.1
SOQ49837.1 NWSH01000895 PCG73653.1 AGBW02011602 OWR46443.1 NWSH01008552 PCG62504.1 JTDY01003429 KOB69569.1 RSAL01000030 RVE51714.1 DS231816 EDS37551.1 DS232267 EDS38787.1 GFDL01010319 JAV24726.1 GFDL01009252 JAV25793.1 AAAB01008980 GGFM01005481 MBW26232.1 APCN01005264 AXCN02001866 GAKP01011642 JAC47310.1 GDHF01006504 JAI45810.1 GBXI01014028 JAD00264.1 DS235134 EEB12407.1 GGFK01009389 MBW42710.1 GGFK01009404 MBW42725.1 EAA13959.5 ADMH02002079 ETN59455.1 GGFJ01007627 MBW56768.1 GL449347 EFN82982.1 GAMC01004018 JAC02538.1 CVRI01000037 CRK93241.1 CH477974 EAT34690.1 CH478060 EAT34149.1 UFQS01001355 UFQT01001355 SSX10497.1 SSX30183.1 UFQT01000874 SSX27761.1 JXUM01119439 JXUM01119440 JXUM01119441 JXUM01119442 JXUM01119443 KQ566290 KXJ70258.1 GFDG01000415 JAV18384.1 GFDG01000416 JAV18383.1 JXJN01002686 EZ423092 ADD19368.1 CP012523 ALC37928.1 CH933807 EDW13053.2 CH940649 KRF81991.1 UFQS01004235 UFQT01004235 SSX16266.1 SSX35587.1 EDW65035.2 JRES01000668 KNC29414.1 OUUW01000004 SPP79119.1 CH902620 EDV30881.1 AAZX01003293 KPU73097.1 CH916368 EDW03461.1 CH379060 EAL33813.1 EU310340 ABY55448.1 SPP79120.1 KQ971363 EFA07837.2 KA649383 AFP64012.1 KA645901 AFP60530.1 AXCM01000224 CM000157 EDW87191.1 EU310337 EU310338 EU310339 EU310341 ABY55445.1 ABY55446.1 ABY55447.1 ABY55449.1 EU670477 ACD56183.1 CH963920 EDW77664.2 EU670475 EU670476 EU670478 CM000361 CM002910 ACD56181.1 ACD56182.1 ACD56184.1 EDX03271.1 KMY87383.1 CH954177 EDV57282.2 GEDC01012856 GEDC01001009 JAS24442.1 JAS36289.1 JR042514 AEY59630.1 EU670516 CH480829 ACD56185.1 EDW45450.1 AE014134 AGB92384.1 AJ000516 AY095092 AY118303 GGFJ01008133 MBW57274.1 JXUM01112104 KQ565562 KXJ70883.1 KQ976511 KYM82449.1 GECU01019787 GECU01010408 GECU01007454 GECU01004744 JAS87919.1 JAS97298.1 JAT00253.1 JAT02963.1 PYGN01000551 PSN44389.1 GEZM01058957 JAV71603.1 GEZM01058958 JAV71602.1 GFXV01000665 MBW12470.1 GEZM01058956 JAV71604.1 GL769036 EFZ10818.1 GL438030 EFN69658.1 QOIP01000010 RLU17385.1 GBHO01000341 GBRD01000894 JAG43263.1 JAG64927.1 ABLF02021049 GDHC01012626 JAQ06003.1 RLU17382.1 KK853018 KDR12428.1 LBMM01001627 KMQ95982.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000037510
+ More
UP000283053 UP000002320 UP000076407 UP000075840 UP000075886 UP000075920 UP000009046 UP000007062 UP000076408 UP000000673 UP000008237 UP000075900 UP000183832 UP000008820 UP000069940 UP000249989 UP000092445 UP000095300 UP000092460 UP000078200 UP000091820 UP000092553 UP000009192 UP000008792 UP000037069 UP000268350 UP000192221 UP000092443 UP000007801 UP000002358 UP000075882 UP000001070 UP000001819 UP000007266 UP000095301 UP000075883 UP000192223 UP000002282 UP000007798 UP000000304 UP000008711 UP000001292 UP000000803 UP000078540 UP000245037 UP000000311 UP000075884 UP000279307 UP000075901 UP000007819 UP000027135 UP000036403 UP000075881
UP000283053 UP000002320 UP000076407 UP000075840 UP000075886 UP000075920 UP000009046 UP000007062 UP000076408 UP000000673 UP000008237 UP000075900 UP000183832 UP000008820 UP000069940 UP000249989 UP000092445 UP000095300 UP000092460 UP000078200 UP000091820 UP000092553 UP000009192 UP000008792 UP000037069 UP000268350 UP000192221 UP000092443 UP000007801 UP000002358 UP000075882 UP000001070 UP000001819 UP000007266 UP000095301 UP000075883 UP000192223 UP000002282 UP000007798 UP000000304 UP000008711 UP000001292 UP000000803 UP000078540 UP000245037 UP000000311 UP000075884 UP000279307 UP000075901 UP000007819 UP000027135 UP000036403 UP000075881
Interpro
CDD
ProteinModelPortal
H9J6X1
A0A194QL80
A0A194Q1W8
A0A2H1W9W4
A0A2A4JQD2
A0A212EY73
+ More
A0A2A4ITD1 A0A0L7L2Q8 A0A3S2M599 B0VZX9 B0X1Q9 A0A1Q3FAX7 A0A1Q3FE02 A0A1S4H3F5 A0A182X7A8 A0A2M3ZCK3 A0A182I0T4 A0A182Q612 A0A034VZL3 A0A182VSV4 A0A0K8W4M4 A0A0A1WMK6 E0VGA1 A0A2M4APK9 A0A2M4APK7 Q7PME1 A0A182YHF7 W5J9D0 A0A2M4BVJ2 E2BMU7 W8BTN0 A0A182RAC0 A0A1J1I4D1 Q16K76 Q16IQ5 A0A336MMY0 A0A336MEG2 A0A182H8W0 A0A1B0A5U1 A0A1I8Q8J6 A0A1L8EIA2 A0A1L8EIG7 A0A1B0AS70 D3TNU0 A0A1A9VH29 A0A1A9X0Q8 A0A0M4DZD8 B4KF70 A0A0Q9WIQ8 A0A336N3V9 B4LTY2 A0A0L0CD77 A0A3B0JZK8 A0A1W4V2Q3 A0A1A9Y0D2 B3MM66 K7JF39 A0A182KJV4 A0A0P8XUU7 B4JDU1 Q29M81 B0FE52 A0A3B0JUG6 D6WZ04 T1PN51 T1PAW0 A0A182M175 A0A1W4WGQ0 B4P2Y4 B0FE49 B2ZI86 B4N0Q1 B2ZI84 B3N895 A0A1B6EEB3 V9IEZ5 B2ZIC5 M9PBM0 O18391 A0A2M4BW91 A0A182H5N0 A0A195BE09 A0A1B6JMD8 A0A2P8YJF8 A0A1Y1LDA6 A0A1Y1LD67 A0A2H8TEJ1 A0A1Y1LDA2 E9J8P2 E2AA78 A0A182N8T6 A0A3L8DA60 A0A182SKB6 A0A0A9ZHH3 J9K888 A0A146LCD4 A0A3L8DB58 A0A067QTY7 A0A0J7KZJ8 A0A182KA99
A0A2A4ITD1 A0A0L7L2Q8 A0A3S2M599 B0VZX9 B0X1Q9 A0A1Q3FAX7 A0A1Q3FE02 A0A1S4H3F5 A0A182X7A8 A0A2M3ZCK3 A0A182I0T4 A0A182Q612 A0A034VZL3 A0A182VSV4 A0A0K8W4M4 A0A0A1WMK6 E0VGA1 A0A2M4APK9 A0A2M4APK7 Q7PME1 A0A182YHF7 W5J9D0 A0A2M4BVJ2 E2BMU7 W8BTN0 A0A182RAC0 A0A1J1I4D1 Q16K76 Q16IQ5 A0A336MMY0 A0A336MEG2 A0A182H8W0 A0A1B0A5U1 A0A1I8Q8J6 A0A1L8EIA2 A0A1L8EIG7 A0A1B0AS70 D3TNU0 A0A1A9VH29 A0A1A9X0Q8 A0A0M4DZD8 B4KF70 A0A0Q9WIQ8 A0A336N3V9 B4LTY2 A0A0L0CD77 A0A3B0JZK8 A0A1W4V2Q3 A0A1A9Y0D2 B3MM66 K7JF39 A0A182KJV4 A0A0P8XUU7 B4JDU1 Q29M81 B0FE52 A0A3B0JUG6 D6WZ04 T1PN51 T1PAW0 A0A182M175 A0A1W4WGQ0 B4P2Y4 B0FE49 B2ZI86 B4N0Q1 B2ZI84 B3N895 A0A1B6EEB3 V9IEZ5 B2ZIC5 M9PBM0 O18391 A0A2M4BW91 A0A182H5N0 A0A195BE09 A0A1B6JMD8 A0A2P8YJF8 A0A1Y1LDA6 A0A1Y1LD67 A0A2H8TEJ1 A0A1Y1LDA2 E9J8P2 E2AA78 A0A182N8T6 A0A3L8DA60 A0A182SKB6 A0A0A9ZHH3 J9K888 A0A146LCD4 A0A3L8DB58 A0A067QTY7 A0A0J7KZJ8 A0A182KA99
PDB
3FBW
E-value=1.26018e-08,
Score=141
Ontologies
GO
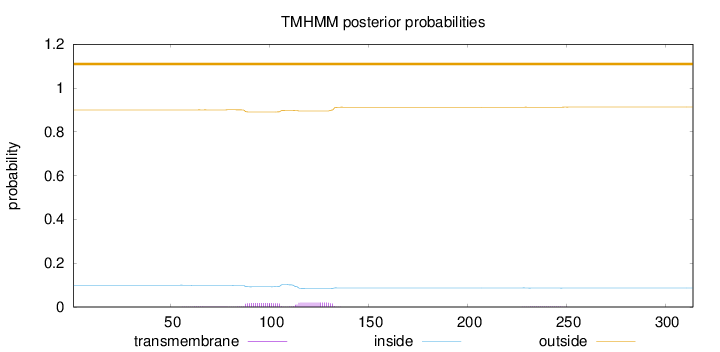
Topology
Length:
314
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.753029999999999
Exp number, first 60 AAs:
0.00175
Total prob of N-in:
0.09989
outside
1 - 314
Population Genetic Test Statistics
Pi
176.417576
Theta
168.729127
Tajima's D
0.581735
CLR
1.025551
CSRT
0.542922853857307
Interpretation
Uncertain
