Pre Gene Modal
BGIBMGA005269
Annotation
PREDICTED:_NFX1-type_zinc_finger-containing_protein_1-like_isoform_X1_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.875
Sequence
CDS
ATGGATAAACCAACAATGCGGATAGATTGGTTTGATGGTACCGTAGTAGAAGATTCCAGATCAATAGCAACAACGTCGCAATTTGCTGACGAGGAAGAAGACATAGATGTTGCAAAAAAACCTACACATAAACAAACAAACACAGAAAAGAAACCAATTGGCTTCAAAAGGCTACTCGATATATCATTATTGAAACCACCCATTTTAACATTAGAAGTCTCTCATAGACCTGGTTTTTGGAATCTTCTAGACGAGGAAGAGCTAAAGGGCGATTTTATAGTTTTAATCGTAAAAATATTGGCTTCCATTTACAAATCTTTGAGAAGTGGCGAGAATAGTAAAATTGTTACAATGCTCAGGACACGATTGAGAAATTCGAAGTTTTTATTGAATTTGAAAAACTACATCACTGATCTACCAGCTGTTCGTATAGTAGAAAAGAGATTGAATACACACATGTGGGATGATGCCGAGGGATTTTTTATTGAAATGGTTGAATTGTGTGAAGGAATCCTTAATTTTGGTGGGAACAGTGAAGAATATTTACAAAACATTTTTGAGTTGTTAGAAGCTACTGAAATTAGTGCTTTAGGAGTTATGGAAGAGCATTCTGAAAGGTTTAGTAATGAACTGTTTGACAGAATCGAAAAAATTAAGATAAGGTTTGCTGAAGAACTTAGTTCAAAGGGTCAGACTATAACAAATGAGACAAGGATAAATGAAGACCCGAACAGTTTCAGAAATCTAAACATATTCCCGACTCAACGAGACCTACTGAGCACTGATCATATTAATTTAAAACCTAACATAATCAACGGAGCGTACCCAAATGTGGAGCATTATCTGGATGTTCAGTTCAAATTGTTAAGGGAAGATTACTTTGGACCTTTAAGAGATGGAATAAGTTGAATACATCCAAAAGTTCGTATAATCCGTAACTATGTGACAAACAACAAAATAGGGTACCTAGTCGATATAGCATGGAATGAGCGATACAACCAAAGTACAATCAACAACACAACACTAAAGTACACCAAGCAACTGATGTTTGGGTCACTTTTACTGTTCACAAATGATAATTTTGAAACGATATTGTGCGCAACTGTACTGGACTCTAGTTATAGTTTATTGGCTGAAGGCTATATTGCTGTAGCCTTTCAAAATCCAGTATCCAATAAAATATTCTCAGAAGCATATTTAATGGTTGAAAGCGAAGTATTTTTTGAGCCTTACCATAGAGTGCTTAAAGTTTTACAAAATTCAAGACCTGACGAGCTTCCCATGAAGAAATACATTGTGGATGTGGACAATGAAACTCAACCACCAAGATATTTGAACTCTGATACGCGATACGCCATAACCGAACCAGGGTGTACTGAAGAAATACAATTCCCAGTCTTGCGACCAGACCTATGGCCAGATGAATCAATAGGCCTAGACGAATCTCAGTTAAATGCATACAAATTCGCTTTGACCAAAGAGTTCGCCGTCATCCAAGGTCCTCCTGGTACCGGAAAAACGTTTTTAGGGGTCAAAATTGCTTCAACGCTACTCAGAAATTTATCCCTAGAAGGCACACCGATGTTGATTATTTGTTATACAAATCACGCCCTTGATCAATTCTTGGAGGGATTGCTCTCTGTGACCCAAAGTATAGTCAGATTAGGAAGCCAAAGTAAAAGTAAGGTTTTGGAGCGGTATAGCTTACACAACGCCAGGATGGGGGTCAAATCTAAGTATTCATATTTGTATGGGACTAAGAGGGCGGAAATAGAAAGGGTTTTTAAAGAAATGTGTGAAGTTCAGAGCGAGATTGAGTTATGCGAAAGAGAGGTGCTCTCATATAAATGCATTAGACCATTCCTGAAGATTGATGGTAAGAGTTATGAATTGAAGAGTTCAAAAGGTGATTCCGTATTGGATTGGCTGTTTGGTTGTGACAATGGAGCTTTTGAACCGGACACAAGTGACGATTGGGAAAATTTAGACGAATCTAGCACAGATATACTAGAAAGTGTATTTTCAGAACATTATGCTTTAAAGGAAATAGATTCAATGTTAAACAGCATAAAATATGTTCAAGATATAACTGATGATGCAGAGGAAAGAAAGCAAATGGTGGATAAGTTTAAAGCACAGATTGATAAAGTTAAAAGTCGATTGGATTGTTTTAAGAAATACCGTACTCTATACCAGATTTACAATGAACCAAAATTAACCATAGACAAAGTTGAAGATCTATACTGTTTGAAATTAGAACAAAGATGGCAACTTTATTACAAAACAGTTGGATGTGTTAAAAGTAACTTACTGTCGAAAATGAATACGCTTTTGGAAAAGCACAAGACGTTGAATCAGGAGTTGGAACAGGTGACGACGTTGACTGATGCCGCCGTAATGGAGAAGGTCCGGGTGGTGGGCGCGACCACCAGCGCGGCGGCCCGGAGACGCGACCTGCTGACCACGCTGCGCCAGTCCGTAGTGATAATTGAAGAAGCAGCCGAAGTCTTAGAGGCTCACATCGTCGCTTCTCTGACCAACGACTGTCAGCACCTGATACTGATAGGTGACCACAAGCAACTTCGTCCTACGGCCGCTAACTATATCCTAGCGAAGCGATACAACCTGGAGGTATCTCTGTTCGAACGAATGATTAGGAACGGTGTACACGCTCGAGTGCTCACAGTGCAGCGCCGCATGAGGCCGCGTGTCGCCGAGCTGCTGGTGCCGACCGTGTACCCTGCCCTAGACTCGCACCCGGACGTGTACAACTACCCGGACGTCAGGGGGATGGCCGATAATCTATACTTCTTCACCCACGAAGTCCTAGAAGATACCGAGGGTTTGGAAGGGAGCTGGAGTCACCGAAATACCTTTGAAGCCAGATGGTGCGTGGTCTTGGCGAATTATTTACGACAAATGAAATATTTGGCTCACGAAGTGACTATACTGTCTACTTATATGGAACAAGTGTCATTAATAAAGCAGTTGAGCTTAAAGTACAATTCGCTACGCGATATAAAGATAACGGCCGTCGATAATTACCAAGGCGAAGAGAGCAGGATAGTGATATTGTCTCTTGTGAGGAGTAACAAGGACGGCAAGATTGGCTTCCTGAATGCCACCAATAGGATCTGCGTGGCTCTATCTAGAGCTAGAGAAGGTTTCTACATATTTGGCAATATGAATTTATTAAAGACTGCGAATAATGTGTGGAGCTCGATCAATGAAAAGCTGATAAAACAAAACGCTATCGGCAATAAATTAACATTGGTGTGTAAACATCATGCGAAACGTTTGGAAGTGAGGACCGTGAACGAATTTGAAAAATACATTTCTGGAACGTGCCCGATGAATTGTGGAAAGAAATAA
Protein
MDKPTMRIDWFDGTVVEDSRSIATTSQFADEEEDIDVAKKPTHKQTNTEKKPIGFKRLLDISLLKPPILTLEVSHRPGFWNLLDEEELKGDFIVLIVKILASIYKSLRSGENSKIVTMLRTRLRNSKFLLNLKNYITDLPAVRIVEKRLNTHMWDDAEGFFIEMVELCEGILNFGGNSEEYLQNIFELLEATEISALGVMEEHSERFSNELFDRIEKIKIRFAEELSSKGQTITNETRINEDPNSFRNLNIFPTQRDLLSTDHINLKPNIINGAYPNVEHYLDVQFKLLREDYFGPLRDGISXIHPKVRIIRNYVTNNKIGYLVDIAWNERYNQSTINNTTLKYTKQLMFGSLLLFTNDNFETILCATVLDSSYSLLAEGYIAVAFQNPVSNKIFSEAYLMVESEVFFEPYHRVLKVLQNSRPDELPMKKYIVDVDNETQPPRYLNSDTRYAITEPGCTEEIQFPVLRPDLWPDESIGLDESQLNAYKFALTKEFAVIQGPPGTGKTFLGVKIASTLLRNLSLEGTPMLIICYTNHALDQFLEGLLSVTQSIVRLGSQSKSKVLERYSLHNARMGVKSKYSYLYGTKRAEIERVFKEMCEVQSEIELCEREVLSYKCIRPFLKIDGKSYELKSSKGDSVLDWLFGCDNGAFEPDTSDDWENLDESSTDILESVFSEHYALKEIDSMLNSIKYVQDITDDAEERKQMVDKFKAQIDKVKSRLDCFKKYRTLYQIYNEPKLTIDKVEDLYCLKLEQRWQLYYKTVGCVKSNLLSKMNTLLEKHKTLNQELEQVTTLTDAAVMEKVRVVGATTSAAARRRDLLTTLRQSVVIIEEAAEVLEAHIVASLTNDCQHLILIGDHKQLRPTAANYILAKRYNLEVSLFERMIRNGVHARVLTVQRRMRPRVAELLVPTVYPALDSHPDVYNYPDVRGMADNLYFFTHEVLEDTEGLEGSWSHRNTFEARWCVVLANYLRQMKYLAHEVTILSTYMEQVSLIKQLSLKYNSLRDIKITAVDNYQGEESRIVILSLVRSNKDGKIGFLNATNRICVALSRAREGFYIFGNMNLLKTANNVWSSINEKLIKQNAIGNKLTLVCKHHAKRLEVRTVNEFEKYISGTCPMNCGKK
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
H9J6X7
A0A2A4JNT7
A0A2H1W9U5
A0A3S2NXS6
A0A194Q8I4
A0A212EYC3
+ More
A0A0L7L283 A0A194RGS3 A0A3S2NYC3 A0A194RH26 A0A2H1WHA2 A0A194PWK9 A0A1Y1KDY9 A0A2J7RCT3 A0A1B6E714 A0A1B6E310 A0A1B6CRR7 D6WH23 A0A2J7REG5 A0A067R9K1 A0A1B6LYJ5 A0A1B6CDC7 A0A1B6D0I0 A0A1B6GP87 A0A1B6H0L9 A0A1B6D8Z8 A0A2S2NW97 A0A1B6JN51 A0A1B6J8F4 A0A1B6CBK5 A0A1B6HVM9 A0A1B6KDN9 A0A146VJF9 A0A1S3DJM9 A0A3Q4GFY5 K7J4A3 A0A232F3L2 A0A3Q2PP61 A0A146VJ96 A0A0P7V5H7 U3IUP4 K7J4A2 A0A1W5A5W2 A0A3B5K2P6 A0A3B4G171 A0A1B6CUY4 A0A1S3H6T7 A0A087TRX9 A0A2I0M261 A0A3P8QKM2 A0A091I9B9 C3ZBY9 A0A091FZT0 A0A3B3XAC4 A0A3Q2V7V5 A0A3P9BAD9 A0A087X8A5 A0A2P6LDQ5 H3CNN6 A0A093P2P4 A0A087R2K2 A0A1V4KKU4 A0A1S3P7Q2 A0A094KNI0 A0A060WYZ5 A0A3P8U2C2 A0A091WEC2 A0A091M0F8 K7J4A1 A0A3B4ZTX9 K7G9L0 A0A099ZSV9 A0A3Q3LED1 A0A2H8TVB5 A0A3M6UCA2 T2M3H7 M7B7Z0 W4YK75 V4AYN5 W4XSS0 T1FY24 A0A2T7NBV5 W4YS82 W4Z6I2 A0A3L8S995 A0A372RFZ8 A0A2N1N3K2 A0A2I1EWR6 A0A2N0RQU9 A0A015KM05 A0A2N0PEP5 U9SLB0 W4ZDB4 A0A2I1GF88 A0A397SMD0 V4ANR5 R7U8Y9
A0A0L7L283 A0A194RGS3 A0A3S2NYC3 A0A194RH26 A0A2H1WHA2 A0A194PWK9 A0A1Y1KDY9 A0A2J7RCT3 A0A1B6E714 A0A1B6E310 A0A1B6CRR7 D6WH23 A0A2J7REG5 A0A067R9K1 A0A1B6LYJ5 A0A1B6CDC7 A0A1B6D0I0 A0A1B6GP87 A0A1B6H0L9 A0A1B6D8Z8 A0A2S2NW97 A0A1B6JN51 A0A1B6J8F4 A0A1B6CBK5 A0A1B6HVM9 A0A1B6KDN9 A0A146VJF9 A0A1S3DJM9 A0A3Q4GFY5 K7J4A3 A0A232F3L2 A0A3Q2PP61 A0A146VJ96 A0A0P7V5H7 U3IUP4 K7J4A2 A0A1W5A5W2 A0A3B5K2P6 A0A3B4G171 A0A1B6CUY4 A0A1S3H6T7 A0A087TRX9 A0A2I0M261 A0A3P8QKM2 A0A091I9B9 C3ZBY9 A0A091FZT0 A0A3B3XAC4 A0A3Q2V7V5 A0A3P9BAD9 A0A087X8A5 A0A2P6LDQ5 H3CNN6 A0A093P2P4 A0A087R2K2 A0A1V4KKU4 A0A1S3P7Q2 A0A094KNI0 A0A060WYZ5 A0A3P8U2C2 A0A091WEC2 A0A091M0F8 K7J4A1 A0A3B4ZTX9 K7G9L0 A0A099ZSV9 A0A3Q3LED1 A0A2H8TVB5 A0A3M6UCA2 T2M3H7 M7B7Z0 W4YK75 V4AYN5 W4XSS0 T1FY24 A0A2T7NBV5 W4YS82 W4Z6I2 A0A3L8S995 A0A372RFZ8 A0A2N1N3K2 A0A2I1EWR6 A0A2N0RQU9 A0A015KM05 A0A2N0PEP5 U9SLB0 W4ZDB4 A0A2I1GF88 A0A397SMD0 V4ANR5 R7U8Y9
Pubmed
EMBL
BABH01028334
BABH01028335
NWSH01000895
PCG73651.1
ODYU01007234
SOQ49838.1
+ More
RSAL01000030 RVE51716.1 KQ459302 KPJ01847.1 AGBW02011602 OWR46444.1 JTDY01003429 KOB69567.1 KQ460211 KPJ16525.1 RSAL01000025 RVE52130.1 KQ460397 KPJ15236.1 ODYU01008646 SOQ52407.1 KQ459589 KPI97707.1 GEZM01088890 JAV57845.1 NEVH01005883 PNF38643.1 GEDC01025993 GEDC01003586 JAS11305.1 JAS33712.1 GEDC01004975 JAS32323.1 GEDC01021385 JAS15913.1 KQ971321 EFA00122.1 NEVH01005006 PNF39219.1 KK852655 KDR19284.1 GEBQ01011287 JAT28690.1 GEDC01025857 GEDC01025145 JAS11441.1 JAS12153.1 GEDC01018110 JAS19188.1 GECZ01005540 JAS64229.1 GECZ01001543 JAS68226.1 GEDC01015131 JAS22167.1 GGMR01008816 MBY21435.1 GECU01007038 JAT00669.1 GECU01012252 JAS95454.1 GEDC01026474 JAS10824.1 GECU01035096 GECU01029016 JAS72610.1 JAS78690.1 GEBQ01030399 GEBQ01016845 JAT09578.1 JAT23132.1 GCES01068928 JAR17395.1 AAZX01003253 NNAY01001131 OXU25033.1 GCES01068927 JAR17396.1 JARO02000941 KPP76956.1 ADON01112229 ADON01112230 GEDC01020153 JAS17145.1 KK116477 KFM67868.1 AKCR02000047 PKK23763.1 KL218292 KFP03950.1 GG666604 EEN49930.1 KL447510 KFO74299.1 AYCK01000328 MWRG01000218 PRD36703.1 KL225346 KFW71273.1 KL226064 KFM07706.1 LSYS01002950 OPJ85064.1 KL351094 KFZ58957.1 FR904817 CDQ72212.1 KK735429 KFR14057.1 KK513965 KFP64109.1 AGCU01006266 AGCU01006267 AGCU01006268 AGCU01006269 AGCU01006270 AGCU01006271 AGCU01006272 KL896797 KGL83860.1 GFXV01006410 MBW18215.1 RCHS01001807 RMX51300.1 HAAD01000606 CDG66838.1 KB564564 EMP28273.1 AAGJ04048993 KB201205 ESO98811.1 AAGJ04001507 AMQM01000597 KB096324 ESO06284.1 PZQS01000014 PVD18632.1 AAGJ04128395 QUSF01000037 RLV98769.1 QKKE01000086 RGB38984.1 LLXL01000837 PKK68482.1 LLXK01000979 PKY26566.1 LLXH01000521 PKC65690.1 JEMT01017251 EXX68564.1 LLXJ01000894 PKC05288.1 KI301740 BDIQ01000044 AUPC02000399 ERZ94797.1 GBC17823.1 POG60277.1 AAGJ04001505 LLXI01000373 PKY45278.1 QKYT01000543 RIA83764.1 ESO98812.1 AMQN01008687 KB303737 ELU02825.1
RSAL01000030 RVE51716.1 KQ459302 KPJ01847.1 AGBW02011602 OWR46444.1 JTDY01003429 KOB69567.1 KQ460211 KPJ16525.1 RSAL01000025 RVE52130.1 KQ460397 KPJ15236.1 ODYU01008646 SOQ52407.1 KQ459589 KPI97707.1 GEZM01088890 JAV57845.1 NEVH01005883 PNF38643.1 GEDC01025993 GEDC01003586 JAS11305.1 JAS33712.1 GEDC01004975 JAS32323.1 GEDC01021385 JAS15913.1 KQ971321 EFA00122.1 NEVH01005006 PNF39219.1 KK852655 KDR19284.1 GEBQ01011287 JAT28690.1 GEDC01025857 GEDC01025145 JAS11441.1 JAS12153.1 GEDC01018110 JAS19188.1 GECZ01005540 JAS64229.1 GECZ01001543 JAS68226.1 GEDC01015131 JAS22167.1 GGMR01008816 MBY21435.1 GECU01007038 JAT00669.1 GECU01012252 JAS95454.1 GEDC01026474 JAS10824.1 GECU01035096 GECU01029016 JAS72610.1 JAS78690.1 GEBQ01030399 GEBQ01016845 JAT09578.1 JAT23132.1 GCES01068928 JAR17395.1 AAZX01003253 NNAY01001131 OXU25033.1 GCES01068927 JAR17396.1 JARO02000941 KPP76956.1 ADON01112229 ADON01112230 GEDC01020153 JAS17145.1 KK116477 KFM67868.1 AKCR02000047 PKK23763.1 KL218292 KFP03950.1 GG666604 EEN49930.1 KL447510 KFO74299.1 AYCK01000328 MWRG01000218 PRD36703.1 KL225346 KFW71273.1 KL226064 KFM07706.1 LSYS01002950 OPJ85064.1 KL351094 KFZ58957.1 FR904817 CDQ72212.1 KK735429 KFR14057.1 KK513965 KFP64109.1 AGCU01006266 AGCU01006267 AGCU01006268 AGCU01006269 AGCU01006270 AGCU01006271 AGCU01006272 KL896797 KGL83860.1 GFXV01006410 MBW18215.1 RCHS01001807 RMX51300.1 HAAD01000606 CDG66838.1 KB564564 EMP28273.1 AAGJ04048993 KB201205 ESO98811.1 AAGJ04001507 AMQM01000597 KB096324 ESO06284.1 PZQS01000014 PVD18632.1 AAGJ04128395 QUSF01000037 RLV98769.1 QKKE01000086 RGB38984.1 LLXL01000837 PKK68482.1 LLXK01000979 PKY26566.1 LLXH01000521 PKC65690.1 JEMT01017251 EXX68564.1 LLXJ01000894 PKC05288.1 KI301740 BDIQ01000044 AUPC02000399 ERZ94797.1 GBC17823.1 POG60277.1 AAGJ04001505 LLXI01000373 PKY45278.1 QKYT01000543 RIA83764.1 ESO98812.1 AMQN01008687 KB303737 ELU02825.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000007151
UP000037510
+ More
UP000053240 UP000235965 UP000007266 UP000027135 UP000079169 UP000261580 UP000002358 UP000215335 UP000265000 UP000034805 UP000016666 UP000192224 UP000005226 UP000261460 UP000085678 UP000054359 UP000053872 UP000265100 UP000054308 UP000001554 UP000053760 UP000261480 UP000264840 UP000265160 UP000028760 UP000007303 UP000054081 UP000053286 UP000190648 UP000087266 UP000193380 UP000265080 UP000053605 UP000261400 UP000007267 UP000053641 UP000261640 UP000275408 UP000031443 UP000007110 UP000030746 UP000015101 UP000245119 UP000276834 UP000263633 UP000233469 UP000232688 UP000022910 UP000232722 UP000018888 UP000236242 UP000234323 UP000265703 UP000014760
UP000053240 UP000235965 UP000007266 UP000027135 UP000079169 UP000261580 UP000002358 UP000215335 UP000265000 UP000034805 UP000016666 UP000192224 UP000005226 UP000261460 UP000085678 UP000054359 UP000053872 UP000265100 UP000054308 UP000001554 UP000053760 UP000261480 UP000264840 UP000265160 UP000028760 UP000007303 UP000054081 UP000053286 UP000190648 UP000087266 UP000193380 UP000265080 UP000053605 UP000261400 UP000007267 UP000053641 UP000261640 UP000275408 UP000031443 UP000007110 UP000030746 UP000015101 UP000245119 UP000276834 UP000263633 UP000233469 UP000232688 UP000022910 UP000232722 UP000018888 UP000236242 UP000234323 UP000265703 UP000014760
PRIDE
Pfam
Interpro
IPR027417
P-loop_NTPase
+ More
IPR041677 DNA2/NAM7_AAA_11
IPR041679 DNA2/NAM7-like_AAA
IPR000967 Znf_NFX1
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR033116 TRYPSIN_SER
IPR003593 AAA+_ATPase
IPR016024 ARM-type_fold
IPR013087 Znf_C2H2_type
IPR014013 Helic_SF1/SF2_ATP-bd_DinG/Rad3
IPR036860 SH2_dom_sf
IPR015424 PyrdxlP-dep_Trfase
IPR005135 Endo/exonuclease/phosphatase
IPR015421 PyrdxlP-dep_Trfase_major
IPR000980 SH2
IPR000192 Aminotrans_V_dom
IPR036691 Endo/exonu/phosph_ase_sf
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR000300 IPPc
IPR014001 Helicase_ATP-bd
IPR041677 DNA2/NAM7_AAA_11
IPR041679 DNA2/NAM7-like_AAA
IPR000967 Znf_NFX1
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR033116 TRYPSIN_SER
IPR003593 AAA+_ATPase
IPR016024 ARM-type_fold
IPR013087 Znf_C2H2_type
IPR014013 Helic_SF1/SF2_ATP-bd_DinG/Rad3
IPR036860 SH2_dom_sf
IPR015424 PyrdxlP-dep_Trfase
IPR005135 Endo/exonuclease/phosphatase
IPR015421 PyrdxlP-dep_Trfase_major
IPR000980 SH2
IPR000192 Aminotrans_V_dom
IPR036691 Endo/exonu/phosph_ase_sf
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR000300 IPPc
IPR014001 Helicase_ATP-bd
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9J6X7
A0A2A4JNT7
A0A2H1W9U5
A0A3S2NXS6
A0A194Q8I4
A0A212EYC3
+ More
A0A0L7L283 A0A194RGS3 A0A3S2NYC3 A0A194RH26 A0A2H1WHA2 A0A194PWK9 A0A1Y1KDY9 A0A2J7RCT3 A0A1B6E714 A0A1B6E310 A0A1B6CRR7 D6WH23 A0A2J7REG5 A0A067R9K1 A0A1B6LYJ5 A0A1B6CDC7 A0A1B6D0I0 A0A1B6GP87 A0A1B6H0L9 A0A1B6D8Z8 A0A2S2NW97 A0A1B6JN51 A0A1B6J8F4 A0A1B6CBK5 A0A1B6HVM9 A0A1B6KDN9 A0A146VJF9 A0A1S3DJM9 A0A3Q4GFY5 K7J4A3 A0A232F3L2 A0A3Q2PP61 A0A146VJ96 A0A0P7V5H7 U3IUP4 K7J4A2 A0A1W5A5W2 A0A3B5K2P6 A0A3B4G171 A0A1B6CUY4 A0A1S3H6T7 A0A087TRX9 A0A2I0M261 A0A3P8QKM2 A0A091I9B9 C3ZBY9 A0A091FZT0 A0A3B3XAC4 A0A3Q2V7V5 A0A3P9BAD9 A0A087X8A5 A0A2P6LDQ5 H3CNN6 A0A093P2P4 A0A087R2K2 A0A1V4KKU4 A0A1S3P7Q2 A0A094KNI0 A0A060WYZ5 A0A3P8U2C2 A0A091WEC2 A0A091M0F8 K7J4A1 A0A3B4ZTX9 K7G9L0 A0A099ZSV9 A0A3Q3LED1 A0A2H8TVB5 A0A3M6UCA2 T2M3H7 M7B7Z0 W4YK75 V4AYN5 W4XSS0 T1FY24 A0A2T7NBV5 W4YS82 W4Z6I2 A0A3L8S995 A0A372RFZ8 A0A2N1N3K2 A0A2I1EWR6 A0A2N0RQU9 A0A015KM05 A0A2N0PEP5 U9SLB0 W4ZDB4 A0A2I1GF88 A0A397SMD0 V4ANR5 R7U8Y9
A0A0L7L283 A0A194RGS3 A0A3S2NYC3 A0A194RH26 A0A2H1WHA2 A0A194PWK9 A0A1Y1KDY9 A0A2J7RCT3 A0A1B6E714 A0A1B6E310 A0A1B6CRR7 D6WH23 A0A2J7REG5 A0A067R9K1 A0A1B6LYJ5 A0A1B6CDC7 A0A1B6D0I0 A0A1B6GP87 A0A1B6H0L9 A0A1B6D8Z8 A0A2S2NW97 A0A1B6JN51 A0A1B6J8F4 A0A1B6CBK5 A0A1B6HVM9 A0A1B6KDN9 A0A146VJF9 A0A1S3DJM9 A0A3Q4GFY5 K7J4A3 A0A232F3L2 A0A3Q2PP61 A0A146VJ96 A0A0P7V5H7 U3IUP4 K7J4A2 A0A1W5A5W2 A0A3B5K2P6 A0A3B4G171 A0A1B6CUY4 A0A1S3H6T7 A0A087TRX9 A0A2I0M261 A0A3P8QKM2 A0A091I9B9 C3ZBY9 A0A091FZT0 A0A3B3XAC4 A0A3Q2V7V5 A0A3P9BAD9 A0A087X8A5 A0A2P6LDQ5 H3CNN6 A0A093P2P4 A0A087R2K2 A0A1V4KKU4 A0A1S3P7Q2 A0A094KNI0 A0A060WYZ5 A0A3P8U2C2 A0A091WEC2 A0A091M0F8 K7J4A1 A0A3B4ZTX9 K7G9L0 A0A099ZSV9 A0A3Q3LED1 A0A2H8TVB5 A0A3M6UCA2 T2M3H7 M7B7Z0 W4YK75 V4AYN5 W4XSS0 T1FY24 A0A2T7NBV5 W4YS82 W4Z6I2 A0A3L8S995 A0A372RFZ8 A0A2N1N3K2 A0A2I1EWR6 A0A2N0RQU9 A0A015KM05 A0A2N0PEP5 U9SLB0 W4ZDB4 A0A2I1GF88 A0A397SMD0 V4ANR5 R7U8Y9
PDB
6QDV
E-value=2.27497e-21,
Score=257
Ontologies
GO
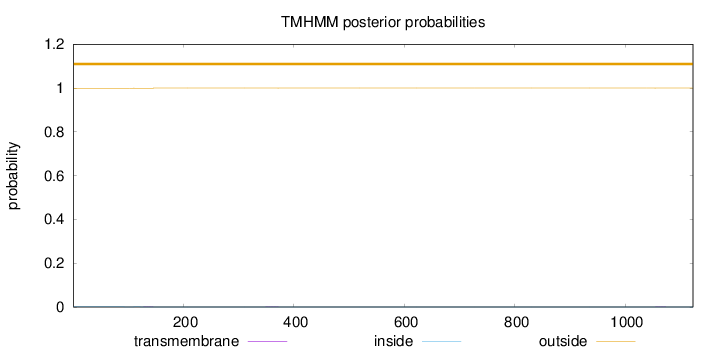
Topology
Length:
1123
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.021
Exp number, first 60 AAs:
0.0005
Total prob of N-in:
0.00073
outside
1 - 1123
Population Genetic Test Statistics
Pi
202.526801
Theta
168.130114
Tajima's D
1.595488
CLR
0.78236
CSRT
0.808409579521024
Interpretation
Uncertain
