Gene
KWMTBOMO04339
Pre Gene Modal
BGIBMGA005266
Annotation
cytochrome_P450_CYP49A1_[Helicoverpa_armigera]
Location in the cell
Mitochondrial Reliability : 3.572
Sequence
CDS
ATGGCGAAGTCAACAGTTGTGATTCGTCAATCATTACTCCAACGACAATGTCGTCGCCACATCGCTGGTTCGTCGTCCAGTCGCACTTCAACATCTCCTCAGCGACGGAACGCAGCCGCAACAGCATCAGCCACGGCAACCTGCTTGAAACCATTCAACGAGATCCCTGGACCCATGGCTTTGCCCATGTTGAGACACTCGGCTCATATCCTACCGAAAATCGGTAATTTCCACCATACTGTAGGTTTGGATGTACTGGAAAACCTTCGTAAGAAATATGGAGATCTAGTGCGACTGTCGAAAGCCACGAGGACTCGTCCCGTACTATACGTATTTCACCCTGAGATGATGAGAGAGGTGTATGAGAGCGGTCAAACGGAGCCACCACATTTCGATCGCTCGCCACTGTGTCATCACAGGAAAGGCGTCGGTAGCCTCTGTCCTATGCACAATGATGAAACAAAGGCCGTGTGGAGTGGCCTTCGATTGCTGCTACAAGAGGAAGCGATCTTGAAGAACTACGATGCTGCGTTCGATGACATCGCTACTGACGTCACCAGGAGACTTGAGGAACTGCGGTGCAAAGGGAATGTCCTTAACGAGGAATTGGAAACGGAGATATATAGATGGGCCCTCGAGACTGTGGGGATGATGTTGTTTGGCATCAGACTCGGCTGTCTGGACGCTCGGACAAAATCAGGAGCCGACTTGTCCCCAACAGAAGAACCCGTCGAAGGTTTATCTCCGGCCGAGAGGCTGGTCCGGTGTAGCCGGCAGATCTCTGAGGGCGCCTTCTTGGTGCGAAGTGAGGAAACTTTGAAGACCGAGTCCCAAACATTCAACGATGCCTTGAAGTCATTCGACCGACATCTGAGCTTAACAGAGCACTTCCTAATCAAAGCGATCCATACGATGACCTCGTGTATTACGAGACCGGAACAAGTACTACTCGATAAATTGAGGCCTCTGGAGAGAAGACTACTTCCCTTGGCGTCGGATATGCTACTGGCGGCTGTCGACCCTCTAGCCCAGACCGCCCTAACGATGTTCTACCAGCTCTCCATCCACGCGGCCCACCAGCAGCGCGCCCACGACGAGGTGCTGTGGTGCAGCGCGTCCCGGGCGGCCGGCGCTGCGCCCCCCGCGGCCCCCGCGCGCCACTCGGTCCCCGCCGCGTCCCCCGCGCCCCCCGCCCCGCAGTACGTGTCGGCGTGCGCGCGGGAGACGCTCCGCCTGCAGCCCCCCACCGGGGGGGTCGTCAGGCGGAGCGTCGACAGCTTGGTGCTCGATGGATTTGAAATACCTGCCGGGGTAGACATCGTTTTAGCGCACGGATTAACGAGCAAGGAAGAAAAACAGTGGGGTAGAGCTAAAGCCTTTGTACCTGAACGATGGTGTGACGATGGCTGGCAGCCTCTGAAGGCATCCAGGGCTCATCACCTAGCCTCCATGCCCTTCGGCGAAAAGTGCCCGGGGACAGGCATCTCTATAAAGATGCTCACATCTCTGGCTGCAAGAATCACCGAGAGATATAGGATAGAGTGGCATGGACCGGCTCCCGCACTGACTCTACATGGAGTCAACAAAATACAAAGACCTTATTATTTTGTGCTACAGAATGCTTCGTGA
Protein
MAKSTVVIRQSLLQRQCRRHIAGSSSSRTSTSPQRRNAAATASATATCLKPFNEIPGPMALPMLRHSAHILPKIGNFHHTVGLDVLENLRKKYGDLVRLSKATRTRPVLYVFHPEMMREVYESGQTEPPHFDRSPLCHHRKGVGSLCPMHNDETKAVWSGLRLLLQEEAILKNYDAAFDDIATDVTRRLEELRCKGNVLNEELETEIYRWALETVGMMLFGIRLGCLDARTKSGADLSPTEEPVEGLSPAERLVRCSRQISEGAFLVRSEETLKTESQTFNDALKSFDRHLSLTEHFLIKAIHTMTSCITRPEQVLLDKLRPLERRLLPLASDMLLAAVDPLAQTALTMFYQLSIHAAHQQRAHDEVLWCSASRAAGAAPPAAPARHSVPAASPAPPAPQYVSACARETLRLQPPTGGVVRRSVDSLVLDGFEIPAGVDIVLAHGLTSKEEKQWGRAKAFVPERWCDDGWQPLKASRAHHLASMPFGEKCPGTGISIKMLTSLAARITERYRIEWHGPAPALTLHGVNKIQRPYYFVLQNAS
Summary
Similarity
Belongs to the cytochrome P450 family.
Uniprot
L0N6M5
L0N7A1
A0A248QG97
A0A068EVJ5
A0A286MXP1
A0A194QA25
+ More
A0A1V0D9I2 A0A2A4J4E6 A0A0L7LQ97 L0N6M0 L0N5L2 J7FIT7 D2JLK5 A0A248QEI8 A0A286QUH6 D5L0N7 A0A212ERS8 H9IVJ7 A0A194QSH9 A0A2A4JB57 A0A0C5C533 A0A194PNU6 A0A068ETR9 A0A1E1WBR0 A0A1E1W6Z4 L0N731 D5L0N6 H9IVJ6 A0A068EWM5 A0A194PD37 D5L0N5 A0A248QEI9 A0A1V0D9G5 J7FJF8 A0A194PCV8 A0A212EU53 A0A1E1WJG1 A0A194QYD6 A0A286MXN7 A0A3S2PF19 A0A0C5C1L2 A0A0C5C1H5 A0A0L7LAT5 K7J4S2 A0A1V0D9G9 A0A1E1WN58 A0A182H3M9 A0A212F2K7 D2A103 A0A195BNU5 A0A151JR78 A0A023ETJ1 E9IWZ0 A0A232FNV9 A0A195FQU6 A0A151WM01 A0A346II03 A0A195CUB6 E0VDR0 A0A2J7RNN8 A0A182QKV0 A0A182PLU9 A0A026WVH3 A0A0M4H594 A0A182S4M1 A0A1E1WAC2 L0N6I8 A0A0K0LB65 A0A182Y248 A0A182XML5 A0A182VDU1 A0A182UA43 A0A182L361 Q7PNM9 A0A182HWH4 A0A0L0BP26 A0A182J2S6 A0A0M4ETM1 A0A1E1W847 B4LKE7 B4N662 Q17JN3 B0WTS8 A0A182FHV4 B4KN43
A0A1V0D9I2 A0A2A4J4E6 A0A0L7LQ97 L0N6M0 L0N5L2 J7FIT7 D2JLK5 A0A248QEI8 A0A286QUH6 D5L0N7 A0A212ERS8 H9IVJ7 A0A194QSH9 A0A2A4JB57 A0A0C5C533 A0A194PNU6 A0A068ETR9 A0A1E1WBR0 A0A1E1W6Z4 L0N731 D5L0N6 H9IVJ6 A0A068EWM5 A0A194PD37 D5L0N5 A0A248QEI9 A0A1V0D9G5 J7FJF8 A0A194PCV8 A0A212EU53 A0A1E1WJG1 A0A194QYD6 A0A286MXN7 A0A3S2PF19 A0A0C5C1L2 A0A0C5C1H5 A0A0L7LAT5 K7J4S2 A0A1V0D9G9 A0A1E1WN58 A0A182H3M9 A0A212F2K7 D2A103 A0A195BNU5 A0A151JR78 A0A023ETJ1 E9IWZ0 A0A232FNV9 A0A195FQU6 A0A151WM01 A0A346II03 A0A195CUB6 E0VDR0 A0A2J7RNN8 A0A182QKV0 A0A182PLU9 A0A026WVH3 A0A0M4H594 A0A182S4M1 A0A1E1WAC2 L0N6I8 A0A0K0LB65 A0A182Y248 A0A182XML5 A0A182VDU1 A0A182UA43 A0A182L361 Q7PNM9 A0A182HWH4 A0A0L0BP26 A0A182J2S6 A0A0M4ETM1 A0A1E1W847 B4LKE7 B4N662 Q17JN3 B0WTS8 A0A182FHV4 B4KN43
Pubmed
EMBL
AK343213
AK343214
BAM73896.1
BAM73897.1
AK289308
BAM73835.1
+ More
KX443483 ASO98058.1 KM016721 AID54873.1 MF684359 ASX93993.1 KQ459302 KPJ01845.1 KY212095 ARA91659.1 NWSH01003063 PCG67017.1 JTDY01000334 KOB77653.1 AK343201 BAM73887.1 AK343196 AK343202 BAM73885.1 BAM73888.1 JX310099 AFP20610.1 GQ915321 ACZ97415.1 KX443480 ASO98055.1 KX443481 ASO98056.1 GU731542 ADE05592.1 AGBW02012935 OWR44198.1 BABH01001231 BABH01001232 KQ461154 KPJ08442.1 NWSH01002099 PCG69201.1 KP001115 AJN91160.1 KQ459599 KPI94419.1 KM016710 KZ149946 AID54862.1 PZC76746.1 GDQN01010473 GDQN01006630 JAT80581.1 JAT84424.1 GDQN01008311 JAT82743.1 AK289309 BAM73836.1 GU731541 ADE05591.1 BABH01001230 KM016709 AID54861.1 KQ459606 KPI91147.1 GU731540 ADE05590.1 KX443479 ASO98054.1 KY212081 ARA91645.1 JX310098 AFP20609.1 KPI91146.1 AGBW02012491 OWR44987.1 GDQN01003905 JAT87149.1 KQ460947 KPJ10553.1 MF684355 ASX93989.1 RSAL01000062 RVE49588.1 KP001114 AJN91159.1 KP001116 AJN91161.1 JTDY01001897 KOB72593.1 AAZX01005602 KY212080 ARA91644.1 GDQN01002676 JAT88378.1 JXUM01024875 JXUM01074800 JXUM01074801 KQ562852 KQ560704 KXJ75006.1 KXJ81216.1 AGBW02010730 OWR47956.1 KQ971338 EFA02807.1 KQ976435 KYM87027.1 KQ978615 KYN29857.1 GAPW01000938 JAC12660.1 GL766616 EFZ14919.1 NNAY01000010 OXU32057.1 KQ981382 KYN42279.1 KQ982959 KYQ48725.1 MH138217 AXP17158.1 KQ977279 KYN04117.1 DS235083 EEB11516.1 NEVH01002150 PNF42438.1 AXCN02000515 AXCN02000516 KK107085 QOIP01000008 EZA60040.1 RLU19440.1 KR012815 ALD15890.1 GDQN01007130 JAT83924.1 AK289310 BAM73837.1 KM216993 AIW79956.1 AAAB01008960 EAA11902.5 APCN01001491 JRES01001578 KNC21835.1 CP012524 ALC40607.1 GDQN01007911 JAT83143.1 CH940648 EDW61738.1 CH964154 EDW79851.2 CH477232 EAT46807.2 DS232091 EDS34542.1 CH933808 EDW08870.1 KRG04326.1 KRG04327.1
KX443483 ASO98058.1 KM016721 AID54873.1 MF684359 ASX93993.1 KQ459302 KPJ01845.1 KY212095 ARA91659.1 NWSH01003063 PCG67017.1 JTDY01000334 KOB77653.1 AK343201 BAM73887.1 AK343196 AK343202 BAM73885.1 BAM73888.1 JX310099 AFP20610.1 GQ915321 ACZ97415.1 KX443480 ASO98055.1 KX443481 ASO98056.1 GU731542 ADE05592.1 AGBW02012935 OWR44198.1 BABH01001231 BABH01001232 KQ461154 KPJ08442.1 NWSH01002099 PCG69201.1 KP001115 AJN91160.1 KQ459599 KPI94419.1 KM016710 KZ149946 AID54862.1 PZC76746.1 GDQN01010473 GDQN01006630 JAT80581.1 JAT84424.1 GDQN01008311 JAT82743.1 AK289309 BAM73836.1 GU731541 ADE05591.1 BABH01001230 KM016709 AID54861.1 KQ459606 KPI91147.1 GU731540 ADE05590.1 KX443479 ASO98054.1 KY212081 ARA91645.1 JX310098 AFP20609.1 KPI91146.1 AGBW02012491 OWR44987.1 GDQN01003905 JAT87149.1 KQ460947 KPJ10553.1 MF684355 ASX93989.1 RSAL01000062 RVE49588.1 KP001114 AJN91159.1 KP001116 AJN91161.1 JTDY01001897 KOB72593.1 AAZX01005602 KY212080 ARA91644.1 GDQN01002676 JAT88378.1 JXUM01024875 JXUM01074800 JXUM01074801 KQ562852 KQ560704 KXJ75006.1 KXJ81216.1 AGBW02010730 OWR47956.1 KQ971338 EFA02807.1 KQ976435 KYM87027.1 KQ978615 KYN29857.1 GAPW01000938 JAC12660.1 GL766616 EFZ14919.1 NNAY01000010 OXU32057.1 KQ981382 KYN42279.1 KQ982959 KYQ48725.1 MH138217 AXP17158.1 KQ977279 KYN04117.1 DS235083 EEB11516.1 NEVH01002150 PNF42438.1 AXCN02000515 AXCN02000516 KK107085 QOIP01000008 EZA60040.1 RLU19440.1 KR012815 ALD15890.1 GDQN01007130 JAT83924.1 AK289310 BAM73837.1 KM216993 AIW79956.1 AAAB01008960 EAA11902.5 APCN01001491 JRES01001578 KNC21835.1 CP012524 ALC40607.1 GDQN01007911 JAT83143.1 CH940648 EDW61738.1 CH964154 EDW79851.2 CH477232 EAT46807.2 DS232091 EDS34542.1 CH933808 EDW08870.1 KRG04326.1 KRG04327.1
Proteomes
UP000053268
UP000218220
UP000037510
UP000007151
UP000005204
UP000053240
+ More
UP000283053 UP000002358 UP000069940 UP000249989 UP000007266 UP000078540 UP000078492 UP000215335 UP000078541 UP000075809 UP000078542 UP000009046 UP000235965 UP000075886 UP000075885 UP000053097 UP000279307 UP000075900 UP000076408 UP000076407 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000037069 UP000075880 UP000092553 UP000008792 UP000007798 UP000008820 UP000002320 UP000069272 UP000009192
UP000283053 UP000002358 UP000069940 UP000249989 UP000007266 UP000078540 UP000078492 UP000215335 UP000078541 UP000075809 UP000078542 UP000009046 UP000235965 UP000075886 UP000075885 UP000053097 UP000279307 UP000075900 UP000076408 UP000076407 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000037069 UP000075880 UP000092553 UP000008792 UP000007798 UP000008820 UP000002320 UP000069272 UP000009192
Pfam
PF00067 p450
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
L0N6M5
L0N7A1
A0A248QG97
A0A068EVJ5
A0A286MXP1
A0A194QA25
+ More
A0A1V0D9I2 A0A2A4J4E6 A0A0L7LQ97 L0N6M0 L0N5L2 J7FIT7 D2JLK5 A0A248QEI8 A0A286QUH6 D5L0N7 A0A212ERS8 H9IVJ7 A0A194QSH9 A0A2A4JB57 A0A0C5C533 A0A194PNU6 A0A068ETR9 A0A1E1WBR0 A0A1E1W6Z4 L0N731 D5L0N6 H9IVJ6 A0A068EWM5 A0A194PD37 D5L0N5 A0A248QEI9 A0A1V0D9G5 J7FJF8 A0A194PCV8 A0A212EU53 A0A1E1WJG1 A0A194QYD6 A0A286MXN7 A0A3S2PF19 A0A0C5C1L2 A0A0C5C1H5 A0A0L7LAT5 K7J4S2 A0A1V0D9G9 A0A1E1WN58 A0A182H3M9 A0A212F2K7 D2A103 A0A195BNU5 A0A151JR78 A0A023ETJ1 E9IWZ0 A0A232FNV9 A0A195FQU6 A0A151WM01 A0A346II03 A0A195CUB6 E0VDR0 A0A2J7RNN8 A0A182QKV0 A0A182PLU9 A0A026WVH3 A0A0M4H594 A0A182S4M1 A0A1E1WAC2 L0N6I8 A0A0K0LB65 A0A182Y248 A0A182XML5 A0A182VDU1 A0A182UA43 A0A182L361 Q7PNM9 A0A182HWH4 A0A0L0BP26 A0A182J2S6 A0A0M4ETM1 A0A1E1W847 B4LKE7 B4N662 Q17JN3 B0WTS8 A0A182FHV4 B4KN43
A0A1V0D9I2 A0A2A4J4E6 A0A0L7LQ97 L0N6M0 L0N5L2 J7FIT7 D2JLK5 A0A248QEI8 A0A286QUH6 D5L0N7 A0A212ERS8 H9IVJ7 A0A194QSH9 A0A2A4JB57 A0A0C5C533 A0A194PNU6 A0A068ETR9 A0A1E1WBR0 A0A1E1W6Z4 L0N731 D5L0N6 H9IVJ6 A0A068EWM5 A0A194PD37 D5L0N5 A0A248QEI9 A0A1V0D9G5 J7FJF8 A0A194PCV8 A0A212EU53 A0A1E1WJG1 A0A194QYD6 A0A286MXN7 A0A3S2PF19 A0A0C5C1L2 A0A0C5C1H5 A0A0L7LAT5 K7J4S2 A0A1V0D9G9 A0A1E1WN58 A0A182H3M9 A0A212F2K7 D2A103 A0A195BNU5 A0A151JR78 A0A023ETJ1 E9IWZ0 A0A232FNV9 A0A195FQU6 A0A151WM01 A0A346II03 A0A195CUB6 E0VDR0 A0A2J7RNN8 A0A182QKV0 A0A182PLU9 A0A026WVH3 A0A0M4H594 A0A182S4M1 A0A1E1WAC2 L0N6I8 A0A0K0LB65 A0A182Y248 A0A182XML5 A0A182VDU1 A0A182UA43 A0A182L361 Q7PNM9 A0A182HWH4 A0A0L0BP26 A0A182J2S6 A0A0M4ETM1 A0A1E1W847 B4LKE7 B4N662 Q17JN3 B0WTS8 A0A182FHV4 B4KN43
PDB
3EL3
E-value=4.73547e-08,
Score=139
Ontologies
GO
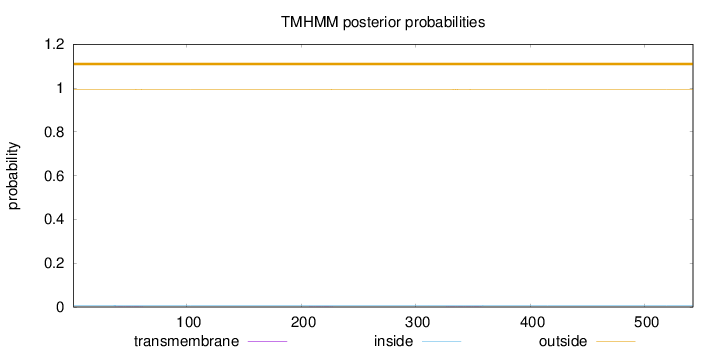
Topology
Length:
542
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0412800000000001
Exp number, first 60 AAs:
0.00921
Total prob of N-in:
0.00635
outside
1 - 542
Population Genetic Test Statistics
Pi
182.700485
Theta
164.914753
Tajima's D
0.582442
CLR
0.434474
CSRT
0.538773061346933
Interpretation
Uncertain
