Gene
KWMTBOMO04323
Pre Gene Modal
BGIBMGA014106
Annotation
PREDICTED:_SUZ_domain-containing_protein_1_[Amyelois_transitella]
Full name
DNA topoisomerase 2
Location in the cell
Nuclear Reliability : 3.651
Sequence
CDS
ATGTACATAAAACAAGGAAGAATGAAGAATCTATGGTTGTCAGTCAACATGGCGGCCCAGGATGAAGTTGACGTATGTGAAAGTTGGGAAGATATGGATGAAATTATTGGTCTGGAACAGAATATCAGGTTAAACAGTGAGAGTCCCACAGAAAACCCGCCAGCTATTTCTCTAAGGGATTGCAAGGTTACTGTTGAAGAGAACGCGTGTATCGGATCGCAGTGCATGATGCCGGAGCCTGCTATTCGCATACTAAAAAGGCCATCATCTTCTGGCAGTGGGCAGTTAAACGGAGAGAATCGTGCCCGACCGGTCAAAACTCTAGAGCAGAGGCAAAAGGAGTATGAACAGGCTCGTCTCAGGATCCTAGGCGAGGCTCGGAGTCCTGAAGATATTATTGATGATAAACTCACCAGGCTACAAGACAAATTACAAGCAAATAATCTTAATGACAGTCAATCAAATAATAAGAAAAATGGTGAAAACGCTAAAAGTAAAGAACGTAAAGTGTTATCTCGGCTTCCACCTGGAGCTACCCCAGCTGGAGTATTTCCATCTCCACCTCCACGAGGAGTGCTTGTTATCCGTACCCCTCGTGGCCCTGATGGCACCAAAGAATATGAGTATACAAGTACAATACTAAAGTTAGTGGTGTTCCAGACTGCAGTCACCTACAGTGATGGTCCGTGCTTAGCTGCAGAGTTTTAG
Protein
MYIKQGRMKNLWLSVNMAAQDEVDVCESWEDMDEIIGLEQNIRLNSESPTENPPAISLRDCKVTVEENACIGSQCMMPEPAIRILKRPSSSGSGQLNGENRARPVKTLEQRQKEYEQARLRILGEARSPEDIIDDKLTRLQDKLQANNLNDSQSNNKKNGENAKSKERKVLSRLPPGATPAGVFPSPPPRGVLVIRTPRGPDGTKEYEYTSTILKLVVFQTAVTYSDGPCLAAEF
Summary
Description
Control of topological states of DNA by transient breakage and subsequent rejoining of DNA strands. Topoisomerase II makes double-strand breaks.
Catalytic Activity
ATP-dependent breakage, passage and rejoining of double-stranded DNA.
Subunit
Homodimer.
Similarity
Belongs to the type II topoisomerase family.
Uniprot
H9JX41
A0A2H1VBC8
A0A212FCL8
A0A194Q042
A0A3S2LUJ4
A0A0L7LSR5
+ More
A0A194RGT8 E0VUS4 A0A2R7WKH0 A0A1W4WPI5 A0A195FM17 A0A026WAA4 A0A1B6LC63 A0A158NF31 A0A151WHV4 A0A1B6H831 A0A3L8DFS9 A0A195E936 A0A195C449 T1I0J3 R4G482 A0A1B6CJC6 E2AE13 A0A087T041 E2C9Y6 A0A1B0D8G1 A0A226E541 A0A0N8CDS3 A0A0N8C719 C4WUF4 A0A0P6CJX5 A0A0P5D4S7 A0A0P4XAX6 A0A2S2P4D7 A0A2I4BDK2 A0A3M7STJ7 A0A087YJB5 A0A3B3UK18
A0A194RGT8 E0VUS4 A0A2R7WKH0 A0A1W4WPI5 A0A195FM17 A0A026WAA4 A0A1B6LC63 A0A158NF31 A0A151WHV4 A0A1B6H831 A0A3L8DFS9 A0A195E936 A0A195C449 T1I0J3 R4G482 A0A1B6CJC6 E2AE13 A0A087T041 E2C9Y6 A0A1B0D8G1 A0A226E541 A0A0N8CDS3 A0A0N8C719 C4WUF4 A0A0P6CJX5 A0A0P5D4S7 A0A0P4XAX6 A0A2S2P4D7 A0A2I4BDK2 A0A3M7STJ7 A0A087YJB5 A0A3B3UK18
EC Number
5.6.2.3
Pubmed
EMBL
BABH01032045
ODYU01001403
SOQ37534.1
AGBW02009166
OWR51482.1
KQ459583
+ More
KPI98663.1 RSAL01000190 RVE44697.1 JTDY01000162 KOB78505.1 KQ460211 KPJ16540.1 DS235793 EEB17130.1 KK854989 PTY20136.1 KQ981490 KYN41371.1 KK107295 EZA52980.1 GEBQ01018727 JAT21250.1 ADTU01013722 KQ983106 KYQ47398.1 GECU01036831 JAS70875.1 QOIP01000009 RLU18759.1 KQ979479 KYN21344.1 KQ978287 KYM95629.1 ACPB03022822 GAHY01001585 JAA75925.1 GEDC01023778 JAS13520.1 GL438827 EFN68246.1 KK112761 KFM58480.1 GL453916 EFN75223.1 AJVK01027392 LNIX01000007 OXA51626.1 GDIP01141663 LRGB01000642 JAL62051.1 KZS17301.1 GDIQ01108507 GDIP01050725 JAL43219.1 JAM52990.1 ABLF02030628 AK341041 BAH71524.1 GDIQ01090213 JAN04524.1 GDIP01161208 JAJ62194.1 GDIP01245263 JAI78138.1 GGMR01011625 MBY24244.1 REGN01000788 RNA39113.1 AYCK01022333
KPI98663.1 RSAL01000190 RVE44697.1 JTDY01000162 KOB78505.1 KQ460211 KPJ16540.1 DS235793 EEB17130.1 KK854989 PTY20136.1 KQ981490 KYN41371.1 KK107295 EZA52980.1 GEBQ01018727 JAT21250.1 ADTU01013722 KQ983106 KYQ47398.1 GECU01036831 JAS70875.1 QOIP01000009 RLU18759.1 KQ979479 KYN21344.1 KQ978287 KYM95629.1 ACPB03022822 GAHY01001585 JAA75925.1 GEDC01023778 JAS13520.1 GL438827 EFN68246.1 KK112761 KFM58480.1 GL453916 EFN75223.1 AJVK01027392 LNIX01000007 OXA51626.1 GDIP01141663 LRGB01000642 JAL62051.1 KZS17301.1 GDIQ01108507 GDIP01050725 JAL43219.1 JAM52990.1 ABLF02030628 AK341041 BAH71524.1 GDIQ01090213 JAN04524.1 GDIP01161208 JAJ62194.1 GDIP01245263 JAI78138.1 GGMR01011625 MBY24244.1 REGN01000788 RNA39113.1 AYCK01022333
Proteomes
UP000005204
UP000007151
UP000053268
UP000283053
UP000037510
UP000053240
+ More
UP000009046 UP000192223 UP000078541 UP000053097 UP000005205 UP000075809 UP000279307 UP000078492 UP000078542 UP000015103 UP000000311 UP000054359 UP000008237 UP000092462 UP000198287 UP000076858 UP000007819 UP000192220 UP000276133 UP000028760 UP000261500
UP000009046 UP000192223 UP000078541 UP000053097 UP000005205 UP000075809 UP000279307 UP000078492 UP000078542 UP000015103 UP000000311 UP000054359 UP000008237 UP000092462 UP000198287 UP000076858 UP000007819 UP000192220 UP000276133 UP000028760 UP000261500
PRIDE
Pfam
Interpro
IPR024771
SUZ
+ More
IPR039228 SZRD1
IPR024642 SUZ-C
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR006171 TOPRIM_domain
IPR013506 Topo_IIA_bsu_dom2
IPR002205 Topo_IIA_A/C
IPR013759 Topo_IIA_B_C
IPR013758 Topo_IIA_A/C_ab
IPR018522 TopoIIA_CS
IPR034157 TOPRIM_TopoII
IPR036890 HATPase_C_sf
IPR013760 Topo_IIA-like_dom_sf
IPR003594 HATPase_C
IPR020568 Ribosomal_S5_D2-typ_fold
IPR013757 Topo_IIA_A_a_sf
IPR001241 Topo_IIA
IPR031660 TOPRIM_C
IPR039228 SZRD1
IPR024642 SUZ-C
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR006171 TOPRIM_domain
IPR013506 Topo_IIA_bsu_dom2
IPR002205 Topo_IIA_A/C
IPR013759 Topo_IIA_B_C
IPR013758 Topo_IIA_A/C_ab
IPR018522 TopoIIA_CS
IPR034157 TOPRIM_TopoII
IPR036890 HATPase_C_sf
IPR013760 Topo_IIA-like_dom_sf
IPR003594 HATPase_C
IPR020568 Ribosomal_S5_D2-typ_fold
IPR013757 Topo_IIA_A_a_sf
IPR001241 Topo_IIA
IPR031660 TOPRIM_C
ProteinModelPortal
H9JX41
A0A2H1VBC8
A0A212FCL8
A0A194Q042
A0A3S2LUJ4
A0A0L7LSR5
+ More
A0A194RGT8 E0VUS4 A0A2R7WKH0 A0A1W4WPI5 A0A195FM17 A0A026WAA4 A0A1B6LC63 A0A158NF31 A0A151WHV4 A0A1B6H831 A0A3L8DFS9 A0A195E936 A0A195C449 T1I0J3 R4G482 A0A1B6CJC6 E2AE13 A0A087T041 E2C9Y6 A0A1B0D8G1 A0A226E541 A0A0N8CDS3 A0A0N8C719 C4WUF4 A0A0P6CJX5 A0A0P5D4S7 A0A0P4XAX6 A0A2S2P4D7 A0A2I4BDK2 A0A3M7STJ7 A0A087YJB5 A0A3B3UK18
A0A194RGT8 E0VUS4 A0A2R7WKH0 A0A1W4WPI5 A0A195FM17 A0A026WAA4 A0A1B6LC63 A0A158NF31 A0A151WHV4 A0A1B6H831 A0A3L8DFS9 A0A195E936 A0A195C449 T1I0J3 R4G482 A0A1B6CJC6 E2AE13 A0A087T041 E2C9Y6 A0A1B0D8G1 A0A226E541 A0A0N8CDS3 A0A0N8C719 C4WUF4 A0A0P6CJX5 A0A0P5D4S7 A0A0P4XAX6 A0A2S2P4D7 A0A2I4BDK2 A0A3M7STJ7 A0A087YJB5 A0A3B3UK18
Ontologies
PANTHER
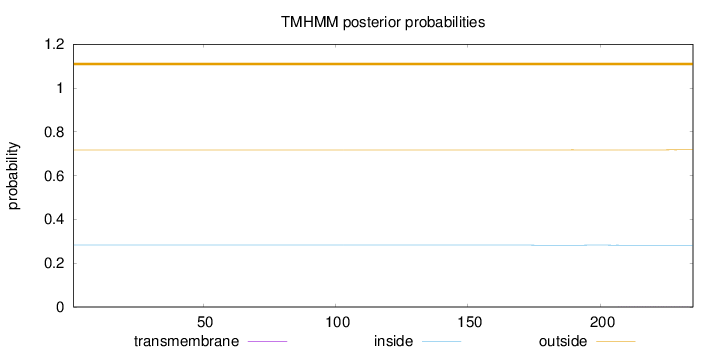
Topology
Length:
235
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0187
Exp number, first 60 AAs:
0
Total prob of N-in:
0.28266
outside
1 - 235
Population Genetic Test Statistics
Pi
311.187238
Theta
224.156807
Tajima's D
1.222622
CLR
0.491087
CSRT
0.719814009299535
Interpretation
Uncertain
