Gene
KWMTBOMO04320
Pre Gene Modal
BGIBMGA012392
Annotation
hypothetical_protein_RR46_02347_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.26 Nuclear Reliability : 1.896
Sequence
CDS
ATGGATAGAATACGGAATGAATATGTTAGAGGAAGTCTGAAAGTGGCACCTGTGACAGAGAAGCTGAGGAGTGCACGTTTGGGATGGTATGGACATGTGATGAGACGAAGTGAAGATGAGGTTGTTAAGAGAGTGTTAACTATGAATGTGGAAGGATTTAGAGGAAGAGGTAGACCTAAGAAGAAATGGATGGATTGTGTGAAAGACGATATGGGTAGGAGGGGAGTGAGCGAAGAAATGGTATATGATAGAAGAGTATGGAAGGAGAAAACATGTTGCGCCGACCCCAGGTGA
Protein
MDRIRNEYVRGSLKVAPVTEKLRSARLGWYGHVMRRSEDEVVKRVLTMNVEGFRGRGRPKKKWMDCVKDDMGRRGVSEEMVYDRRVWKEKTCCADPR
Summary
Uniprot
A0A2A4JJS9
A0A2W1BR99
A0A2A4IZ11
A0A2W1BN03
A0A2W1BM48
A0A194Q6W3
+ More
A0A2W1BIS5 A0A3Q0JQ03 A0A016SZZ3 A0A0C2FK24 A0A183FUR9 A0A3P7YQT3 A0A183GPW8 A0A3P8ETQ0 A0A183F5G0 A0A3P7X1W4 A0A183GBY9 A0A3P8F965 A0A183GB79 A0A3P8EZ99 A0A016UE98 A0A016SDN2 A0A183G681 A0A3P8EH61 A0A016WFX2 A0A016WVF2 A0A183GXF7 A0A3P8EGJ9 A0A183FVH1 A0A3P7YVX3 A0A183F5X8 A0A3P7U6S8 A0A183GPX7 A0A3P8D6Y9 A0A183GDA1 A0A3P8F3X7 A0A016VYQ7 A0A183GPJ0 A0A3P8EHQ4 A0A016TN23 A0A183GWH3 A0A3P8EUL0 A0A016VQL7 A0A183GT84 A0A3P8DS65 A0A183FHI6 A0A3P7Y445 A0A183GTC3 A0A3P8DT03 A0A183F3N4 A0A3P7TGE4 A0A183G9B3 A0A3P8ADM1 A0A183FY09 A0A3P8AX44 A0A0N4YNB7 A0A183FJ73 A0A3P7XZM2 A0A016VNT7 A0A183GBL6 A0A3P8AM59 A0A016UAZ5 A0A183FMV2 A0A3P7XT20 A0A016WLS1 A0A183F9F7 A0A3P7UZH7 A0A183GFE8 A0A3P8FJ37 A0A016U5Z2 A0A016TQG5 A0A016VD78 A0A016VFS4 A0A016VLS9 A0A016VC06 A0A016TKX6 A0A3Q0ISK7 A0A183FB99 A0A3P7VL99 A0A016SIC4 A0A016S9A4 A0A183F3L1 A0A3P7U9P5 A0A016WMJ7 A0A016UG54 A0A183FT77 A0A3P8AAR6 A0A016TK27 A0A016UBL7 A0A183GQD2 A0A3P8HXE7 A0A3B3BCY6 A0A3B3HPR4 A0A183FJ84 A0A3P7YDN4 A0A183G528 A0A3P8E2T0 A0A3P9IVQ2 A0A3P9K1J1
A0A2W1BIS5 A0A3Q0JQ03 A0A016SZZ3 A0A0C2FK24 A0A183FUR9 A0A3P7YQT3 A0A183GPW8 A0A3P8ETQ0 A0A183F5G0 A0A3P7X1W4 A0A183GBY9 A0A3P8F965 A0A183GB79 A0A3P8EZ99 A0A016UE98 A0A016SDN2 A0A183G681 A0A3P8EH61 A0A016WFX2 A0A016WVF2 A0A183GXF7 A0A3P8EGJ9 A0A183FVH1 A0A3P7YVX3 A0A183F5X8 A0A3P7U6S8 A0A183GPX7 A0A3P8D6Y9 A0A183GDA1 A0A3P8F3X7 A0A016VYQ7 A0A183GPJ0 A0A3P8EHQ4 A0A016TN23 A0A183GWH3 A0A3P8EUL0 A0A016VQL7 A0A183GT84 A0A3P8DS65 A0A183FHI6 A0A3P7Y445 A0A183GTC3 A0A3P8DT03 A0A183F3N4 A0A3P7TGE4 A0A183G9B3 A0A3P8ADM1 A0A183FY09 A0A3P8AX44 A0A0N4YNB7 A0A183FJ73 A0A3P7XZM2 A0A016VNT7 A0A183GBL6 A0A3P8AM59 A0A016UAZ5 A0A183FMV2 A0A3P7XT20 A0A016WLS1 A0A183F9F7 A0A3P7UZH7 A0A183GFE8 A0A3P8FJ37 A0A016U5Z2 A0A016TQG5 A0A016VD78 A0A016VFS4 A0A016VLS9 A0A016VC06 A0A016TKX6 A0A3Q0ISK7 A0A183FB99 A0A3P7VL99 A0A016SIC4 A0A016S9A4 A0A183F3L1 A0A3P7U9P5 A0A016WMJ7 A0A016UG54 A0A183FT77 A0A3P8AAR6 A0A016TK27 A0A016UBL7 A0A183GQD2 A0A3P8HXE7 A0A3B3BCY6 A0A3B3HPR4 A0A183FJ84 A0A3P7YDN4 A0A183G528 A0A3P8E2T0 A0A3P9IVQ2 A0A3P9K1J1
EMBL
NWSH01001232
PCG72026.1
KZ150002
PZC75290.1
NWSH01004369
PCG65177.1
+ More
KZ150034 PZC74657.1 KZ150032 PZC74715.1 KQ459581 KPI99130.1 KZ150166 PZC72650.1 JARK01001491 EYB95904.1 KN768094 KIH47114.1 UZAH01027307 VDO90480.1 UZAH01036764 VDP46995.1 UZAH01001522 VDO19785.1 UZAH01031516 VDP15986.1 UZAH01031291 VDP14793.1 JARK01001380 EYC13251.1 JARK01001582 EYB88451.1 UZAH01029847 VDP08183.1 JARK01000336 EYC38182.1 JARK01000107 EYC42998.1 UZAH01043455 VDP63277.1 UZAH01027446 VDO91691.1 UZAH01001784 VDO19994.1 UZAH01036784 VDP47042.1 UZAH01031956 VDP18789.1 JARK01001339 EYC31903.1 UZAH01036580 VDP46188.1 JARK01001494 JARK01001424 EYB95586.1 EYC04409.1 UZAH01041723 VDP60601.1 JARK01001342 EYC29596.1 UZAH01038793 VDP54537.1 UZAH01025623 VDO67507.1 UZAH01038885 VDP54822.1 UZAH01000669 VDO19159.1 UZAH01030740 VDP11902.1 UZAH01027944 VDO96393.1 UYSL01023630 VDL82448.1 UZAH01025799 VDO70679.1 JARK01001343 EYC28702.1 UZAH01031410 VDP15327.1 JARK01001383 EYC12479.1 UZAH01026244 VDO77671.1 JARK01000223 EYC40242.1 UZAH01004968 VDO28364.1 UZAH01032715 VDP23439.1 JARK01001507 JARK01001392 JARK01001372 EYB94492.1 EYC10272.1 EYC15469.1 JARK01001481 JARK01001420 EYB96960.1 EYC04966.1 JARK01001347 EYC25574.1 EYC25573.1 EYC28385.1 JARK01001348 EYC24955.1 JARK01001430 EYC03360.1 UZAH01009099 VDO35114.1 JARK01001560 EYB90061.1 JARK01001602 EYB87210.1 UZAH01000639 VDO19137.1 JARK01000211 EYC40482.1 JARK01001379 EYC13548.1 UZAH01027034 VDO87933.1 EYC03359.1 JARK01001384 EYC12232.1 UZAH01037041 VDP47936.1 UZAH01025803 VDO70747.1 UZAH01029552 VDP06679.1
KZ150034 PZC74657.1 KZ150032 PZC74715.1 KQ459581 KPI99130.1 KZ150166 PZC72650.1 JARK01001491 EYB95904.1 KN768094 KIH47114.1 UZAH01027307 VDO90480.1 UZAH01036764 VDP46995.1 UZAH01001522 VDO19785.1 UZAH01031516 VDP15986.1 UZAH01031291 VDP14793.1 JARK01001380 EYC13251.1 JARK01001582 EYB88451.1 UZAH01029847 VDP08183.1 JARK01000336 EYC38182.1 JARK01000107 EYC42998.1 UZAH01043455 VDP63277.1 UZAH01027446 VDO91691.1 UZAH01001784 VDO19994.1 UZAH01036784 VDP47042.1 UZAH01031956 VDP18789.1 JARK01001339 EYC31903.1 UZAH01036580 VDP46188.1 JARK01001494 JARK01001424 EYB95586.1 EYC04409.1 UZAH01041723 VDP60601.1 JARK01001342 EYC29596.1 UZAH01038793 VDP54537.1 UZAH01025623 VDO67507.1 UZAH01038885 VDP54822.1 UZAH01000669 VDO19159.1 UZAH01030740 VDP11902.1 UZAH01027944 VDO96393.1 UYSL01023630 VDL82448.1 UZAH01025799 VDO70679.1 JARK01001343 EYC28702.1 UZAH01031410 VDP15327.1 JARK01001383 EYC12479.1 UZAH01026244 VDO77671.1 JARK01000223 EYC40242.1 UZAH01004968 VDO28364.1 UZAH01032715 VDP23439.1 JARK01001507 JARK01001392 JARK01001372 EYB94492.1 EYC10272.1 EYC15469.1 JARK01001481 JARK01001420 EYB96960.1 EYC04966.1 JARK01001347 EYC25574.1 EYC25573.1 EYC28385.1 JARK01001348 EYC24955.1 JARK01001430 EYC03360.1 UZAH01009099 VDO35114.1 JARK01001560 EYB90061.1 JARK01001602 EYB87210.1 UZAH01000639 VDO19137.1 JARK01000211 EYC40482.1 JARK01001379 EYC13548.1 UZAH01027034 VDO87933.1 EYC03359.1 JARK01001384 EYC12232.1 UZAH01037041 VDP47936.1 UZAH01025803 VDO70747.1 UZAH01029552 VDP06679.1
Proteomes
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
A0A2A4JJS9
A0A2W1BR99
A0A2A4IZ11
A0A2W1BN03
A0A2W1BM48
A0A194Q6W3
+ More
A0A2W1BIS5 A0A3Q0JQ03 A0A016SZZ3 A0A0C2FK24 A0A183FUR9 A0A3P7YQT3 A0A183GPW8 A0A3P8ETQ0 A0A183F5G0 A0A3P7X1W4 A0A183GBY9 A0A3P8F965 A0A183GB79 A0A3P8EZ99 A0A016UE98 A0A016SDN2 A0A183G681 A0A3P8EH61 A0A016WFX2 A0A016WVF2 A0A183GXF7 A0A3P8EGJ9 A0A183FVH1 A0A3P7YVX3 A0A183F5X8 A0A3P7U6S8 A0A183GPX7 A0A3P8D6Y9 A0A183GDA1 A0A3P8F3X7 A0A016VYQ7 A0A183GPJ0 A0A3P8EHQ4 A0A016TN23 A0A183GWH3 A0A3P8EUL0 A0A016VQL7 A0A183GT84 A0A3P8DS65 A0A183FHI6 A0A3P7Y445 A0A183GTC3 A0A3P8DT03 A0A183F3N4 A0A3P7TGE4 A0A183G9B3 A0A3P8ADM1 A0A183FY09 A0A3P8AX44 A0A0N4YNB7 A0A183FJ73 A0A3P7XZM2 A0A016VNT7 A0A183GBL6 A0A3P8AM59 A0A016UAZ5 A0A183FMV2 A0A3P7XT20 A0A016WLS1 A0A183F9F7 A0A3P7UZH7 A0A183GFE8 A0A3P8FJ37 A0A016U5Z2 A0A016TQG5 A0A016VD78 A0A016VFS4 A0A016VLS9 A0A016VC06 A0A016TKX6 A0A3Q0ISK7 A0A183FB99 A0A3P7VL99 A0A016SIC4 A0A016S9A4 A0A183F3L1 A0A3P7U9P5 A0A016WMJ7 A0A016UG54 A0A183FT77 A0A3P8AAR6 A0A016TK27 A0A016UBL7 A0A183GQD2 A0A3P8HXE7 A0A3B3BCY6 A0A3B3HPR4 A0A183FJ84 A0A3P7YDN4 A0A183G528 A0A3P8E2T0 A0A3P9IVQ2 A0A3P9K1J1
A0A2W1BIS5 A0A3Q0JQ03 A0A016SZZ3 A0A0C2FK24 A0A183FUR9 A0A3P7YQT3 A0A183GPW8 A0A3P8ETQ0 A0A183F5G0 A0A3P7X1W4 A0A183GBY9 A0A3P8F965 A0A183GB79 A0A3P8EZ99 A0A016UE98 A0A016SDN2 A0A183G681 A0A3P8EH61 A0A016WFX2 A0A016WVF2 A0A183GXF7 A0A3P8EGJ9 A0A183FVH1 A0A3P7YVX3 A0A183F5X8 A0A3P7U6S8 A0A183GPX7 A0A3P8D6Y9 A0A183GDA1 A0A3P8F3X7 A0A016VYQ7 A0A183GPJ0 A0A3P8EHQ4 A0A016TN23 A0A183GWH3 A0A3P8EUL0 A0A016VQL7 A0A183GT84 A0A3P8DS65 A0A183FHI6 A0A3P7Y445 A0A183GTC3 A0A3P8DT03 A0A183F3N4 A0A3P7TGE4 A0A183G9B3 A0A3P8ADM1 A0A183FY09 A0A3P8AX44 A0A0N4YNB7 A0A183FJ73 A0A3P7XZM2 A0A016VNT7 A0A183GBL6 A0A3P8AM59 A0A016UAZ5 A0A183FMV2 A0A3P7XT20 A0A016WLS1 A0A183F9F7 A0A3P7UZH7 A0A183GFE8 A0A3P8FJ37 A0A016U5Z2 A0A016TQG5 A0A016VD78 A0A016VFS4 A0A016VLS9 A0A016VC06 A0A016TKX6 A0A3Q0ISK7 A0A183FB99 A0A3P7VL99 A0A016SIC4 A0A016S9A4 A0A183F3L1 A0A3P7U9P5 A0A016WMJ7 A0A016UG54 A0A183FT77 A0A3P8AAR6 A0A016TK27 A0A016UBL7 A0A183GQD2 A0A3P8HXE7 A0A3B3BCY6 A0A3B3HPR4 A0A183FJ84 A0A3P7YDN4 A0A183G528 A0A3P8E2T0 A0A3P9IVQ2 A0A3P9K1J1
Ontologies
GO
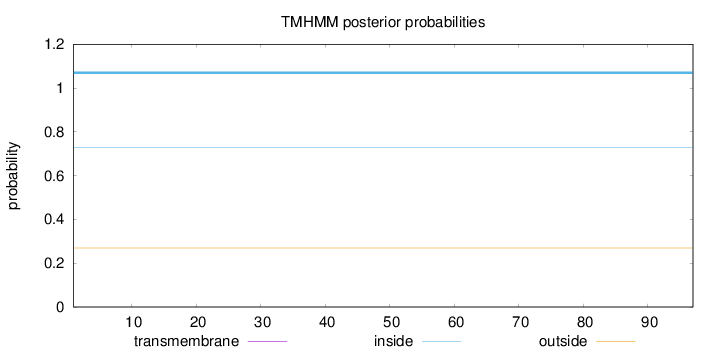
Topology
Length:
97
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.73014
inside
1 - 97
Population Genetic Test Statistics
Pi
246.039396
Theta
125.371698
Tajima's D
2.772133
CLR
0
CSRT
0.96395180240988
Interpretation
Uncertain
