Gene
KWMTBOMO04318
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.131
Sequence
CDS
ATGCATGTGATTAGGAGATGTATGGAAATGGTAGTGCAAGGTAGAGAGGGAAGAGGTCGACCGAAGAAGACATGGATGGAGTGTGTGAATGACGATATGAGAGGGAGAGAGGAGCTAATATCGGTTCACCTCGTGATCGGAGGCGTCCGCGTCGGGGTCGGGGTGACCGGTCTTCGGTACCTCGGCCTCGAACTGGACGGTCGGTGGAACTTCCGCGCTCACTTTGAGAAGTTGGGCCCTCGGCTGATGGCAACGGCCGGCTCTTTGAGCCGGCTCCTTCCAAACGTCGGGGGTCCCGATCAGGTGGTGCGCCGCCTCTACATGGGGGTGGTGCGGTCGATGGCACTATACGGCGCGCCCGTATGGTGCCACGCCCTGACCCGCCAGAACGTCGCCGCGCTGCGACGTCCGCAGCGCGCGATCGCGGTCAGGGCGATCCGAGGATACCGCACCGTCTCCTTCGAGGCGGCGTGCTTGCTCGCCGGGGCGCCACCCTGGGACCTGGAGGCGGAGGCGCTCGCTGCCGATTACCGGTGGCGTAGCGACCTCCGCTCTAGGGGGGAAGGGCGCCCCGGCGAAGGAGTAGTTCGAGCGCGGAGGCTCCAATCTCGGCGGTCCGTGCTGGAGGCGTGGTCTCGCCGCTTGGCGGACCCGTCGGCCGGCCTCCGTACTGTCGAGGCGGTCCGTCCGGTCCTCGCGGACTGGGTGGGCCGCGACCGCGGATGCCTCACCTTCCGGCTGACGCAAATGTTGACCGGCCATGGTTGTTTTGGCCGGTACTTGCACAAGATAGCCGGAAGGGAACCGACGGCGCAGTGCCATCACTGCGCGGACCGCGACGAGGATGACACAGCGGAACACACGCTGGCGCGTTGCTCTGGATTCGACGAGCAACGCGCCGCCCTCGTCGCGGTCATTGGAGAGGACCTCTCGCTGCCGCGCGTCGTGGCTACGATGCTCGGCAGCGATGCGTCCTGGAAGGCGATGCTCGACTTCTGCGAGTCCACCATCTCGCAGAAGGAGGCGGCGGAGCGAGAGAGGGAGAGCTCTTCCCTTTCCGCACCGATCCGTCGCCGTCGAGCCGGGGGCCGGAGGCGGGAATACGCCCGTACGTCCCGGCCCCTGTAG
Protein
MHVIRRCMEMVVQGREGRGRPKKTWMECVNDDMRGREELISVHLVIGGVRVGVGVTGLRYLGLELDGRWNFRAHFEKLGPRLMATAGSLSRLLPNVGGPDQVVRRLYMGVVRSMALYGAPVWCHALTRQNVAALRRPQRAIAVRAIRGYRTVSFEAACLLAGAPPWDLEAEALAADYRWRSDLRSRGEGRPGEGVVRARRLQSRRSVLEAWSRRLADPSAGLRTVEAVRPVLADWVGRDRGCLTFRLTQMLTGHGCFGRYLHKIAGREPTAQCHHCADRDEDDTAEHTLARCSGFDEQRAALVAVIGEDLSLPRVVATMLGSDASWKAMLDFCESTISQKEAAERERESSSLSAPIRRRRAGGRRREYARTSRPL
Summary
Uniprot
Q5KTM7
Q868Q4
A0A2W1BDU5
A0A2W1B333
Q8MY27
Q8MY33
+ More
Q8MY31 Q8MY35 A0A3S2N998 A0A2W1C3J5 A0A0J7N1D9 O01419 A0A0J7N7H9 A0A0J7KCQ3 E2BVR6 A0A0J7KIY5 A0A194R5D3 A0A0J7KS67 A0A0J7N7S8 A0A0J7KIW1 A0A0J7KJG6 A0A0J7KM81 A0A0J7JZ13 A0A3L8DDV9 A0A0J7JZF7 A0A0J7KJH4 A0A3L8D2G3 A0A0J7KQK3 A0A0J7KIM6 A0A0J7KFG9 A0A0J7KLT9 A0A0J7KMU1 A0A0J7K3R6 A0A0J7KPZ3 A0A0J7K3F1 A0A0J7KPR0 A0A0J7NMN5 J9KJG8 A0A3L8DIL1 A0A0J7KYD2 A0A2H8TR36 A0A0J7KRB8 A0A2S2P6S8 A0A2S2NME4 A0A0J7JYP2 A0A0J7NAV8 A0A2S2P557 A0A0J7MX22 A0A0J7KWU5 J9KV04 A0A2S2PDK9 A0A2S2QUS9 A0A2H8TIT6 A0A0A9Z5Y1 A0A0J7KW81 A0A0J7K3R2
Q8MY31 Q8MY35 A0A3S2N998 A0A2W1C3J5 A0A0J7N1D9 O01419 A0A0J7N7H9 A0A0J7KCQ3 E2BVR6 A0A0J7KIY5 A0A194R5D3 A0A0J7KS67 A0A0J7N7S8 A0A0J7KIW1 A0A0J7KJG6 A0A0J7KM81 A0A0J7JZ13 A0A3L8DDV9 A0A0J7JZF7 A0A0J7KJH4 A0A3L8D2G3 A0A0J7KQK3 A0A0J7KIM6 A0A0J7KFG9 A0A0J7KLT9 A0A0J7KMU1 A0A0J7K3R6 A0A0J7KPZ3 A0A0J7K3F1 A0A0J7KPR0 A0A0J7NMN5 J9KJG8 A0A3L8DIL1 A0A0J7KYD2 A0A2H8TR36 A0A0J7KRB8 A0A2S2P6S8 A0A2S2NME4 A0A0J7JYP2 A0A0J7NAV8 A0A2S2P557 A0A0J7MX22 A0A0J7KWU5 J9KV04 A0A2S2PDK9 A0A2S2QUS9 A0A2H8TIT6 A0A0A9Z5Y1 A0A0J7KW81 A0A0J7K3R2
EMBL
AB126050
BAD86650.1
AB090825
BAC57926.1
KZ150308
PZC71477.1
+ More
KZ150386 PZC71062.1 AB078935 BAC06462.1 AB078930 BAC06454.1 AB078931 BAC06456.1 AB078929 BAC06452.1 RSAL01000481 RVE41577.1 KZ149896 PZC78733.1 LBMM01012027 KMQ86470.1 D85594 BAA19776.1 LBMM01008861 KMQ88580.1 LBMM01009494 KMQ88097.1 GL450953 EFN80213.1 LBMM01006954 KMQ90166.1 KQ460685 KPJ12887.1 LBMM01003679 KMQ93242.1 LBMM01008682 KMQ88705.1 LBMM01006725 KMQ90373.1 LBMM01006671 KMQ90407.1 LBMM01005634 KMQ91349.1 LBMM01019839 KMQ83438.1 QOIP01000010 RLU18098.1 LBMM01019838 KMQ83439.1 LBMM01006657 KMQ90417.1 QOIP01000031 RLU14697.1 LBMM01004272 KMQ92618.1 LBMM01007036 KMQ90099.1 LBMM01008349 KMQ88951.1 LBMM01005733 KMQ91252.1 LBMM01005216 KMQ91703.1 LBMM01015018 KMQ84954.1 LBMM01004505 KMQ92346.1 LBMM01015025 KMQ84953.1 LBMM01004634 KMQ92224.1 LBMM01003245 KMQ93755.1 ABLF02041886 QOIP01000007 RLU20280.1 LBMM01002052 KMQ95336.1 GFXV01003903 MBW15708.1 LBMM01003988 KMQ92922.1 GGMR01012534 MBY25153.1 GGMR01005740 MBY18359.1 LBMM01020849 KMQ83202.1 LBMM01007425 KMQ89740.1 GGMR01011974 MBY24593.1 LBMM01014909 KMQ85000.1 LBMM01002480 KMQ94776.1 ABLF02004674 ABLF02004675 ABLF02004676 ABLF02011637 GGMR01014918 MBY27537.1 GGMS01012210 MBY81413.1 GFXV01002242 MBW14047.1 GBHO01002982 JAG40622.1 KMQ94772.1 LBMM01014956 KMQ84982.1
KZ150386 PZC71062.1 AB078935 BAC06462.1 AB078930 BAC06454.1 AB078931 BAC06456.1 AB078929 BAC06452.1 RSAL01000481 RVE41577.1 KZ149896 PZC78733.1 LBMM01012027 KMQ86470.1 D85594 BAA19776.1 LBMM01008861 KMQ88580.1 LBMM01009494 KMQ88097.1 GL450953 EFN80213.1 LBMM01006954 KMQ90166.1 KQ460685 KPJ12887.1 LBMM01003679 KMQ93242.1 LBMM01008682 KMQ88705.1 LBMM01006725 KMQ90373.1 LBMM01006671 KMQ90407.1 LBMM01005634 KMQ91349.1 LBMM01019839 KMQ83438.1 QOIP01000010 RLU18098.1 LBMM01019838 KMQ83439.1 LBMM01006657 KMQ90417.1 QOIP01000031 RLU14697.1 LBMM01004272 KMQ92618.1 LBMM01007036 KMQ90099.1 LBMM01008349 KMQ88951.1 LBMM01005733 KMQ91252.1 LBMM01005216 KMQ91703.1 LBMM01015018 KMQ84954.1 LBMM01004505 KMQ92346.1 LBMM01015025 KMQ84953.1 LBMM01004634 KMQ92224.1 LBMM01003245 KMQ93755.1 ABLF02041886 QOIP01000007 RLU20280.1 LBMM01002052 KMQ95336.1 GFXV01003903 MBW15708.1 LBMM01003988 KMQ92922.1 GGMR01012534 MBY25153.1 GGMR01005740 MBY18359.1 LBMM01020849 KMQ83202.1 LBMM01007425 KMQ89740.1 GGMR01011974 MBY24593.1 LBMM01014909 KMQ85000.1 LBMM01002480 KMQ94776.1 ABLF02004674 ABLF02004675 ABLF02004676 ABLF02011637 GGMR01014918 MBY27537.1 GGMS01012210 MBY81413.1 GFXV01002242 MBW14047.1 GBHO01002982 JAG40622.1 KMQ94772.1 LBMM01014956 KMQ84982.1
Proteomes
Pfam
Interpro
Gene 3D
ProteinModelPortal
Q5KTM7
Q868Q4
A0A2W1BDU5
A0A2W1B333
Q8MY27
Q8MY33
+ More
Q8MY31 Q8MY35 A0A3S2N998 A0A2W1C3J5 A0A0J7N1D9 O01419 A0A0J7N7H9 A0A0J7KCQ3 E2BVR6 A0A0J7KIY5 A0A194R5D3 A0A0J7KS67 A0A0J7N7S8 A0A0J7KIW1 A0A0J7KJG6 A0A0J7KM81 A0A0J7JZ13 A0A3L8DDV9 A0A0J7JZF7 A0A0J7KJH4 A0A3L8D2G3 A0A0J7KQK3 A0A0J7KIM6 A0A0J7KFG9 A0A0J7KLT9 A0A0J7KMU1 A0A0J7K3R6 A0A0J7KPZ3 A0A0J7K3F1 A0A0J7KPR0 A0A0J7NMN5 J9KJG8 A0A3L8DIL1 A0A0J7KYD2 A0A2H8TR36 A0A0J7KRB8 A0A2S2P6S8 A0A2S2NME4 A0A0J7JYP2 A0A0J7NAV8 A0A2S2P557 A0A0J7MX22 A0A0J7KWU5 J9KV04 A0A2S2PDK9 A0A2S2QUS9 A0A2H8TIT6 A0A0A9Z5Y1 A0A0J7KW81 A0A0J7K3R2
Q8MY31 Q8MY35 A0A3S2N998 A0A2W1C3J5 A0A0J7N1D9 O01419 A0A0J7N7H9 A0A0J7KCQ3 E2BVR6 A0A0J7KIY5 A0A194R5D3 A0A0J7KS67 A0A0J7N7S8 A0A0J7KIW1 A0A0J7KJG6 A0A0J7KM81 A0A0J7JZ13 A0A3L8DDV9 A0A0J7JZF7 A0A0J7KJH4 A0A3L8D2G3 A0A0J7KQK3 A0A0J7KIM6 A0A0J7KFG9 A0A0J7KLT9 A0A0J7KMU1 A0A0J7K3R6 A0A0J7KPZ3 A0A0J7K3F1 A0A0J7KPR0 A0A0J7NMN5 J9KJG8 A0A3L8DIL1 A0A0J7KYD2 A0A2H8TR36 A0A0J7KRB8 A0A2S2P6S8 A0A2S2NME4 A0A0J7JYP2 A0A0J7NAV8 A0A2S2P557 A0A0J7MX22 A0A0J7KWU5 J9KV04 A0A2S2PDK9 A0A2S2QUS9 A0A2H8TIT6 A0A0A9Z5Y1 A0A0J7KW81 A0A0J7K3R2
Ontologies
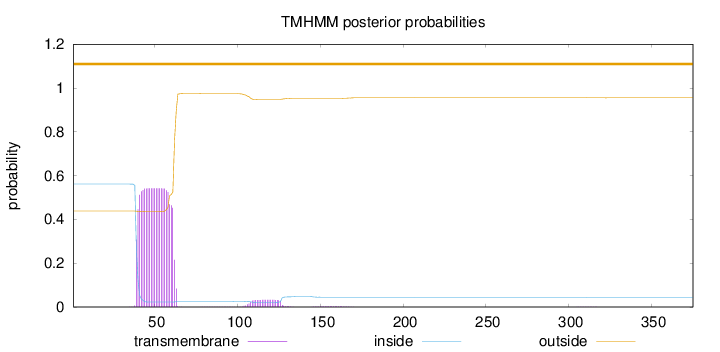
Topology
Length:
375
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
12.84414
Exp number, first 60 AAs:
11.37166
Total prob of N-in:
0.56174
POSSIBLE N-term signal
sequence
outside
1 - 375
Population Genetic Test Statistics
Pi
35.23086
Theta
41.465351
Tajima's D
0.90992
CLR
37.228222
CSRT
0.634868256587171
Interpretation
Uncertain
