Gene
KWMTBOMO04316
Pre Gene Modal
BGIBMGA014109
Annotation
PREDICTED:_rac_GTPase-activating_protein_1-like_[Plutella_xylostella]
Location in the cell
Nuclear Reliability : 2.998
Sequence
CDS
ATGAGTTCAGAAAATTCAGATTCCGGTCCAAGCATCGCGCTCTCCCTTGTAGCCCAATTTGATGACATCAACCGCTTGAATAATGTTTTAAATGATGGAGCCGCAGAAGAATATTTCTTAGCATTCTTAAAGCAAATGGAATGGGTGCGCTGTCAATGGGCAGCAGCCGAGTCCGAGGCTGTTCGCCTTCAAAGTGAGCTTGACGAAGCTCTAAAGACACTAGGCAAGTGGGAATCTAAGTTTGCTCATGTCAGGAAACTGCTTGATGGAGAAAAAAGAGAAAGAAACATAATATTGAAGAAGTACAATGAACTGAGTAAACTACTGGACATGGCCCGTGACCTCTTATTCAATGACAATCGTGCCAAGTTGAATGATGAAACCTTACAGAAGCTTGCATTCCTTAATGGAACTGCTCAACAAGACCACAATGCTAAGCTTAGTGGAGTTCCAGAACTTAATTCTACTGGTACACTACTATCAGAGATGTCATATTCTCGTTCGGAAGATGACCTAGACCTGTCACTGGCTTCGTCTCGACGTTCTTGGGTTCAGCGCGGCGGGTGGCATGCCAGAGAGAACCAGCCCGCCAATAAGAAAAGGCGATCGTCAACTTCCTCTGCTACTAAAACAGTGGAGTTGCAGGGCGGCAAGGTGCTGGCCACCGCTACCACTACACTGACACTGGAACGAGCCCCAACCAACCATGACCTCAAGCCTCCACAGAACTTTCAACCAATATCGGAGAGTAGCGACGAAGCACCACCCCCTCCACGGAAGTCTTCAGCTCGTACATCGGGGGGTGGCGTGACTGCCATCAAAGAGGAGTATACACCCTCAGCACCGCCTCGTACAGAAACAGAGAGCGGGTCGGACACCCCCCGCGCGGCGCCGCAGCGGACCCCCTCCATCATCTCTGCGGCCTACGGCTCGCCCCGCAACCGCGTGCGCCAACACAACTTCGTGGCCAAGAACTTCTACAAGAGGGAGACATGCGGGCCCTGTGGGAAGACCATCAAGTTTGCTAAGATGGGCGTGAAATGCGAGTCGTGCCGCGGGCAGGCGCACCACGAGTGCCGCGCCATGCTGCCGCTGCCCTGCGTGCCGCCGCCCGCGCAGGCGCCGCGACACACGCAGCAGGAGGGCTGTATCTCTGACTACGCGCCCACCACCCCGCCGATGGTGCCGGCCTTGATGGTACATTGTATTAATGAGATCGAGAAACGCGGACTCAGCGAGAGGGGAATCTACAGGATCAGTGCCACAGAGAAGGATGTTAAAAGGCTCAAGGAGCGGTTCCTGCGCGGGCACGGGTCGCCGTGGCTGGCGGGCGAGGACGTGCACGCGGTGTGCGGCTGCGTGAAGGACTTCCTGCGCGGACTGCGCGAGCCGCTGGTGAGCGCGGCGCTGTGGGCCGACTTCATGCAGGCCGCCGCGCTGCCCGACCGCGCCGACGCCGCCGCCGCGCTGCTGCACGCCGTCACGCAGCTGCCGCCGCCCAACCGCGACACGCTCGCCTTCCTCGTGCTGCATCTGCAGAAAGTGGCAGAGAGCCCCGAATGCGAGATGGGCATAGACAATCTGGCTAAAATATTTGGTCCGACCGTCGTCGGGTACGGCATGATGTCGCAAGCCGCGGAGATGTACGCCGCCACAGCGCAGATGTTCAAGGTGATGCAGATAATGCTGTCGCTGCCGAGCGAGTACTGGGCGCAGTGGGCGTGCCCGGCGGACGGCGCGGGCTCGCCGCGGCCCCGGGCCTTCTTCTTTTCGCCGGCCGACACGGGTGTGTCGAGGAAGAAGCGCAACTATTTCCAGTGA
Protein
MSSENSDSGPSIALSLVAQFDDINRLNNVLNDGAAEEYFLAFLKQMEWVRCQWAAAESEAVRLQSELDEALKTLGKWESKFAHVRKLLDGEKRERNIILKKYNELSKLLDMARDLLFNDNRAKLNDETLQKLAFLNGTAQQDHNAKLSGVPELNSTGTLLSEMSYSRSEDDLDLSLASSRRSWVQRGGWHARENQPANKKRRSSTSSATKTVELQGGKVLATATTTLTLERAPTNHDLKPPQNFQPISESSDEAPPPPRKSSARTSGGGVTAIKEEYTPSAPPRTETESGSDTPRAAPQRTPSIISAAYGSPRNRVRQHNFVAKNFYKRETCGPCGKTIKFAKMGVKCESCRGQAHHECRAMLPLPCVPPPAQAPRHTQQEGCISDYAPTTPPMVPALMVHCINEIEKRGLSERGIYRISATEKDVKRLKERFLRGHGSPWLAGEDVHAVCGCVKDFLRGLREPLVSAALWADFMQAAALPDRADAAAALLHAVTQLPPPNRDTLAFLVLHLQKVAESPECEMGIDNLAKIFGPTVVGYGMMSQAAEMYAATAQMFKVMQIMLSLPSEYWAQWACPADGAGSPRPRAFFFSPADTGVSRKKRNYFQ
Summary
Uniprot
A0A2A4J702
S4P8B9
A0A194PZ90
A0A212EHK2
A0A194RGU9
A0A0C9RVA7
+ More
A0A2A3EK46 A0A088A8S3 A0A154PHL2 A0A0J7N4N6 E2AGL2 A0A0M8ZV47 W8BTB2 A0A1I8NZN5 U4UBF4 N6SXR9 A0A0L0C199 A0A3B0J0N5 A0A1W4VG13 D6WE23 A0A1A9YQW3 A0A1A9VW66 A0A0P6EIM3 A0A0P5A6L5 B4J591 A0A164WP98 A0A0P6IEL0 A0A1J1HXT6 A0A1I8MY60 E9I1D9 V9KKT3 A0A0P5QBK3 T1HE66 A0A0P5XWY4 A0A3Q0SBD0 A0A226EHM0 A0A0P5RFX6 A0A0P6FX77 G1PTH8 A0A3B4F1A7 A0A1A8V9D2
A0A2A3EK46 A0A088A8S3 A0A154PHL2 A0A0J7N4N6 E2AGL2 A0A0M8ZV47 W8BTB2 A0A1I8NZN5 U4UBF4 N6SXR9 A0A0L0C199 A0A3B0J0N5 A0A1W4VG13 D6WE23 A0A1A9YQW3 A0A1A9VW66 A0A0P6EIM3 A0A0P5A6L5 B4J591 A0A164WP98 A0A0P6IEL0 A0A1J1HXT6 A0A1I8MY60 E9I1D9 V9KKT3 A0A0P5QBK3 T1HE66 A0A0P5XWY4 A0A3Q0SBD0 A0A226EHM0 A0A0P5RFX6 A0A0P6FX77 G1PTH8 A0A3B4F1A7 A0A1A8V9D2
Pubmed
EMBL
NWSH01002802
PCG67506.1
GAIX01004239
JAA88321.1
KQ459583
KPI98652.1
+ More
AGBW02014869 OWR40966.1 KQ460211 KPJ16550.1 GBYB01012795 JAG82562.1 KZ288230 PBC31632.1 KQ434899 KZC10818.1 LBMM01010208 KMQ87610.1 GL439322 EFN67432.1 KQ435855 KOX70636.1 GAMC01006358 GAMC01006357 JAC00199.1 KB632003 ERL87916.1 APGK01052935 KB741216 ENN72549.1 JRES01001139 KNC25229.1 OUUW01000001 SPP74275.1 KQ971324 EFA01222.2 GDIQ01061952 JAN32785.1 GDIP01203419 JAJ19983.1 CH916367 EDW00717.1 LRGB01001151 KZS13451.1 GDIQ01005675 JAN89062.1 CVRI01000035 CRK92895.1 GL733767 EFX62190.1 JW866272 AFO98789.1 GDIQ01119521 JAL32205.1 ACPB03026840 GDIP01066205 JAM37510.1 LNIX01000003 OXA57112.1 GDIQ01101240 JAL50486.1 GDIQ01052813 JAN41924.1 AAPE02023535 HAEJ01015979 SBS56436.1
AGBW02014869 OWR40966.1 KQ460211 KPJ16550.1 GBYB01012795 JAG82562.1 KZ288230 PBC31632.1 KQ434899 KZC10818.1 LBMM01010208 KMQ87610.1 GL439322 EFN67432.1 KQ435855 KOX70636.1 GAMC01006358 GAMC01006357 JAC00199.1 KB632003 ERL87916.1 APGK01052935 KB741216 ENN72549.1 JRES01001139 KNC25229.1 OUUW01000001 SPP74275.1 KQ971324 EFA01222.2 GDIQ01061952 JAN32785.1 GDIP01203419 JAJ19983.1 CH916367 EDW00717.1 LRGB01001151 KZS13451.1 GDIQ01005675 JAN89062.1 CVRI01000035 CRK92895.1 GL733767 EFX62190.1 JW866272 AFO98789.1 GDIQ01119521 JAL32205.1 ACPB03026840 GDIP01066205 JAM37510.1 LNIX01000003 OXA57112.1 GDIQ01101240 JAL50486.1 GDIQ01052813 JAN41924.1 AAPE02023535 HAEJ01015979 SBS56436.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000053240
UP000242457
UP000005203
+ More
UP000076502 UP000036403 UP000000311 UP000053105 UP000095300 UP000030742 UP000019118 UP000037069 UP000268350 UP000192221 UP000007266 UP000092443 UP000078200 UP000001070 UP000076858 UP000183832 UP000095301 UP000000305 UP000015103 UP000261340 UP000198287 UP000001074 UP000261460
UP000076502 UP000036403 UP000000311 UP000053105 UP000095300 UP000030742 UP000019118 UP000037069 UP000268350 UP000192221 UP000007266 UP000092443 UP000078200 UP000001070 UP000076858 UP000183832 UP000095301 UP000000305 UP000015103 UP000261340 UP000198287 UP000001074 UP000261460
Interpro
SUPFAM
SSF48350
SSF48350
Gene 3D
CDD
ProteinModelPortal
A0A2A4J702
S4P8B9
A0A194PZ90
A0A212EHK2
A0A194RGU9
A0A0C9RVA7
+ More
A0A2A3EK46 A0A088A8S3 A0A154PHL2 A0A0J7N4N6 E2AGL2 A0A0M8ZV47 W8BTB2 A0A1I8NZN5 U4UBF4 N6SXR9 A0A0L0C199 A0A3B0J0N5 A0A1W4VG13 D6WE23 A0A1A9YQW3 A0A1A9VW66 A0A0P6EIM3 A0A0P5A6L5 B4J591 A0A164WP98 A0A0P6IEL0 A0A1J1HXT6 A0A1I8MY60 E9I1D9 V9KKT3 A0A0P5QBK3 T1HE66 A0A0P5XWY4 A0A3Q0SBD0 A0A226EHM0 A0A0P5RFX6 A0A0P6FX77 G1PTH8 A0A3B4F1A7 A0A1A8V9D2
A0A2A3EK46 A0A088A8S3 A0A154PHL2 A0A0J7N4N6 E2AGL2 A0A0M8ZV47 W8BTB2 A0A1I8NZN5 U4UBF4 N6SXR9 A0A0L0C199 A0A3B0J0N5 A0A1W4VG13 D6WE23 A0A1A9YQW3 A0A1A9VW66 A0A0P6EIM3 A0A0P5A6L5 B4J591 A0A164WP98 A0A0P6IEL0 A0A1J1HXT6 A0A1I8MY60 E9I1D9 V9KKT3 A0A0P5QBK3 T1HE66 A0A0P5XWY4 A0A3Q0SBD0 A0A226EHM0 A0A0P5RFX6 A0A0P6FX77 G1PTH8 A0A3B4F1A7 A0A1A8V9D2
PDB
3W6R
E-value=1.56453e-45,
Score=463
Ontologies
KEGG
GO
GO:0046872
GO:0035556
GO:0007165
GO:0090090
GO:0001709
GO:0032465
GO:0042060
GO:0048471
GO:0008283
GO:0055028
GO:0000281
GO:0007525
GO:0005819
GO:0030496
GO:0005634
GO:0070507
GO:0032507
GO:2000736
GO:0048813
GO:0007266
GO:0051988
GO:0005654
GO:0032154
GO:0005547
GO:0019901
GO:0051233
GO:0045995
GO:0000915
GO:0008272
GO:0097149
GO:0005096
GO:0043014
GO:0007405
GO:0043015
GO:0007283
GO:0048487
GO:0032467
GO:0008017
GO:0051256
GO:0031234
GO:0000226
GO:0016757
GO:0006508
GO:0005622
GO:0009113
GO:0005524
GO:0009058
GO:0016742
GO:0003824
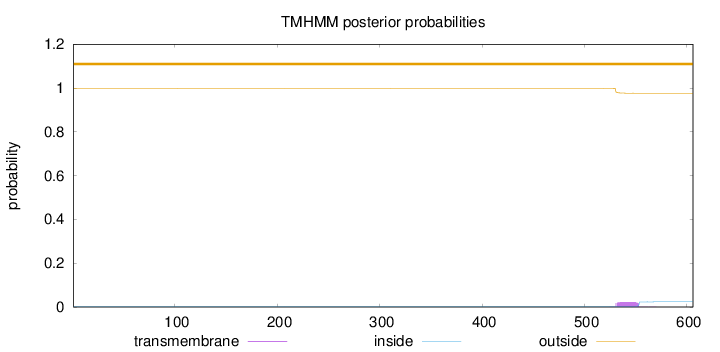
Topology
Length:
606
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.50678
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00065
outside
1 - 606
Population Genetic Test Statistics
Pi
240.986837
Theta
191.77178
Tajima's D
1.134379
CLR
0.384572
CSRT
0.692165391730413
Interpretation
Uncertain
