Gene
KWMTBOMO04305
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_LOC105841899?_partial_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.768 PlasmaMembrane Reliability : 1.687
Sequence
CDS
ATGCTTAACTATTCTGTGCGGACGGCGATACCGGTTCCCGGTCCCCGGTTCCCGGTCGTGCGCGGTGTTCCACAGGGGTCGGTGCTCGGCCCCCTTTTGTGGAATATCGGGTATGACTGGGTGCTGAGAGATGCCCTCCTCCCGGGCCTGAGCGTAATCTGTTACGCGGACGACACGTTGGTCGTGGCCCGGGGGGGAATTTTGCTGAGTCTTGCTACGGCTGGGGTGGCGCATGTCGTCGGCAAAATCAGGAGATTGGGCCTCGACGTGGCGCTGAGTAAATCCGAGGTCATGTGGTTCCACAGGCCCCGGAGAGTGCCACCTGTCGATGCCCATATCGTGGTTGGAGGCGTCCGTATCGGGGTCGGGGTGCAGTTGAAGTACCTCGGCCTCATTCTGGACAGTCGTTGGACCTTCCGTGCTCACTTTCAGAACCTGGTCCCTCGTTTGTTGGGGGTGGCCGGCGCGTTAAGCCGGCTTCTGCCCAACGTCGGGGGGCCTGACCAGGTGACGCGCCGTCTTTATACGGGGATGGTGCGATCAATGGCCCTATACGGGGCGCCCGTGTGGGGCCAGTCCCTGGCCGTGAGTGTGGCGAAGCTGCTGCAACGGCCGCAACGCACCATCGCGGTCAGCGTCATCCGTGGTTATCGCACCATCTCCTTTGAGGCGGCGTGTGTACTGGCTGGGACGCCACCTTGGGTTCTGGAAGCGGAGGCGCTCGCCGCTGACTATAAGTGGCGGGCTGACCTTCGTTCCCGAGGCGTGGCGCGTCCCAGCCCCAGTGTGGTCAGAGCGCGGAGGGCCCAATCTCAGCGGTCCGTGCTGGAGTTATGGTCCAAGCGGCTGGCTAATCCTTCGGCTGGTCGTAAGACCGTCGAGGCGATTCGCCCGGTTCTTGTGGATTGGGTGAATCGTGACAAAGGACGCCTCACTTTCCGGCTCACGCAGGTGTTCACTGGGAATGGTCTTTCGTTCAACTCTGACATACCCCGCCCAGTATTTTCACTCTGTCGTAATGATGCATTTGGGTTTGCAAAAGAAAAACAGTACTACAATACTTTTACGCCGGTAAGAGTAACGCCATCTATCGCCGAATAG
Protein
MLNYSVRTAIPVPGPRFPVVRGVPQGSVLGPLLWNIGYDWVLRDALLPGLSVICYADDTLVVARGGILLSLATAGVAHVVGKIRRLGLDVALSKSEVMWFHRPRRVPPVDAHIVVGGVRIGVGVQLKYLGLILDSRWTFRAHFQNLVPRLLGVAGALSRLLPNVGGPDQVTRRLYTGMVRSMALYGAPVWGQSLAVSVAKLLQRPQRTIAVSVIRGYRTISFEAACVLAGTPPWVLEAEALAADYKWRADLRSRGVARPSPSVVRARRAQSQRSVLELWSKRLANPSAGRKTVEAIRPVLVDWVNRDKGRLTFRLTQVFTGNGLSFNSDIPRPVFSLCRNDAFGFAKEKQYYNTFTPVRVTPSIAE
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
Q868Q4
A0A2W1B333
Q8MY33
Q8MY31
Q8MY35
Q8MY27
+ More
A0A0J7JX58 A0A2W1C3J5 O01419 A0A3S2N998 A0A0J7JWS3 A0A0J7N1D9 A0A194R5D3 A0A0J7KL06 A0A0J7KC11 A0A0J7JZ13 A0A0J7KIY5 A0A0J7KJG6 A0A0J7KM11 A0A0J7JZF7 A0A0J7KYD2 A0A0J7KQK3 A0A0J7KF37 A0A0J7N7S8 A0A0J7KJH4 A0A0J7KLT9 A0A0J7KPR0 A0A0J7JXX4 A0A0J7MYL9 A0A0J7KSR8 A0A0J7KIM6 A0A0J7KTW8 A0A0J7KIW1 A0A0J7KQY4 A0A0J7NJI3 A0A0J7MY07 A0A0J7NAV8 A0A2S2N8I2 A0A0J7NMN5 A0A0J7KF40 A0A0J7K3F1 A0A0J7KGJ6 A0A3L8DDV9 A0A0J7NDI6 A0A3L8D2G3 A0A0J7KRB8 A0A0J7K3R6 A0A0J7KPZ3 A0A0J7KP92 A0A0J7KCZ9 A0A0J7KRB1 A0A0J7MWN5 A0A0J7MY17 A0A0J7KMG5 A0A2S2PBP5 A0A0J7KWU5 A0A0J7KMU1 A0A0A9Z5Y1 A0A0J7K3R2 A0A0J7NFZ0 A0A1B6HLL2 A0A2S2PEZ7 A0A0J7KW81 A0A2S2PIX1 A0A0J7KLK2 A0A2S2PEU2 J9KNG8 A0A0J7K7P2 X1WJY4 J9LJ80 A0A0J7N0R3 A0A0J7KFH2 A0A0A1WPA4 A0A3L8D4C5 A0A2H8THF8 A0A2S2NQQ6 A0A142LX37 A0A139W942
A0A0J7JX58 A0A2W1C3J5 O01419 A0A3S2N998 A0A0J7JWS3 A0A0J7N1D9 A0A194R5D3 A0A0J7KL06 A0A0J7KC11 A0A0J7JZ13 A0A0J7KIY5 A0A0J7KJG6 A0A0J7KM11 A0A0J7JZF7 A0A0J7KYD2 A0A0J7KQK3 A0A0J7KF37 A0A0J7N7S8 A0A0J7KJH4 A0A0J7KLT9 A0A0J7KPR0 A0A0J7JXX4 A0A0J7MYL9 A0A0J7KSR8 A0A0J7KIM6 A0A0J7KTW8 A0A0J7KIW1 A0A0J7KQY4 A0A0J7NJI3 A0A0J7MY07 A0A0J7NAV8 A0A2S2N8I2 A0A0J7NMN5 A0A0J7KF40 A0A0J7K3F1 A0A0J7KGJ6 A0A3L8DDV9 A0A0J7NDI6 A0A3L8D2G3 A0A0J7KRB8 A0A0J7K3R6 A0A0J7KPZ3 A0A0J7KP92 A0A0J7KCZ9 A0A0J7KRB1 A0A0J7MWN5 A0A0J7MY17 A0A0J7KMG5 A0A2S2PBP5 A0A0J7KWU5 A0A0J7KMU1 A0A0A9Z5Y1 A0A0J7K3R2 A0A0J7NFZ0 A0A1B6HLL2 A0A2S2PEZ7 A0A0J7KW81 A0A2S2PIX1 A0A0J7KLK2 A0A2S2PEU2 J9KNG8 A0A0J7K7P2 X1WJY4 J9LJ80 A0A0J7N0R3 A0A0J7KFH2 A0A0A1WPA4 A0A3L8D4C5 A0A2H8THF8 A0A2S2NQQ6 A0A142LX37 A0A139W942
Pubmed
EMBL
AB090825
BAC57926.1
KZ150386
PZC71062.1
AB078930
BAC06454.1
+ More
AB078931 BAC06456.1 AB078929 BAC06452.1 AB078935 BAC06462.1 LBMM01024882 KMQ82501.1 KZ149896 PZC78733.1 D85594 BAA19776.1 RSAL01000481 RVE41577.1 LBMM01024063 KMQ82623.1 LBMM01012027 KMQ86470.1 KQ460685 KPJ12887.1 LBMM01006063 KMQ90967.1 LBMM01009738 KMQ87908.1 LBMM01019839 KMQ83438.1 LBMM01006954 KMQ90166.1 LBMM01006671 KMQ90407.1 LBMM01005707 KMQ91279.1 LBMM01019838 KMQ83439.1 LBMM01002052 KMQ95336.1 LBMM01004272 KMQ92618.1 LBMM01008448 KMQ88867.1 LBMM01008682 KMQ88705.1 LBMM01006657 KMQ90417.1 LBMM01005733 KMQ91252.1 LBMM01004634 KMQ92224.1 LBMM01021647 KMQ83033.1 LBMM01013668 KMQ85535.1 LBMM01003575 KMQ93366.1 LBMM01007036 KMQ90099.1 LBMM01003302 KMQ93689.1 LBMM01006725 KMQ90373.1 LBMM01004262 KMQ92649.1 LBMM01004261 KMQ92650.1 LBMM01014132 KMQ85305.1 LBMM01007425 KMQ89740.1 GGMR01000779 MBY13398.1 LBMM01003245 KMQ93755.1 LBMM01008422 KMQ88894.1 LBMM01015025 KMQ84953.1 LBMM01007659 KMQ89538.1 QOIP01000010 RLU18098.1 LBMM01006496 KMQ90570.1 QOIP01000031 RLU14697.1 LBMM01003988 KMQ92922.1 LBMM01015018 KMQ84954.1 LBMM01004505 KMQ92346.1 LBMM01004697 KMQ92167.1 LBMM01009527 LBMM01009515 KMQ88059.1 KMQ88067.1 LBMM01004138 KMQ92774.1 LBMM01015285 KMQ84870.1 LBMM01014122 KMQ85315.1 LBMM01005465 KMQ91492.1 GGMR01014195 MBY26814.1 LBMM01002480 KMQ94776.1 LBMM01005216 KMQ91703.1 GBHO01002982 JAG40622.1 LBMM01014956 KMQ84982.1 LBMM01005474 KMQ91485.1 GECU01032129 JAS75577.1 GGMR01015159 MBY27778.1 KMQ94772.1 GGMR01016764 MBY29383.1 LBMM01005845 KMQ91162.1 GGMR01015354 MBY27973.1 ABLF02005203 ABLF02041316 LBMM01012322 LBMM01012138 KMQ86282.1 KMQ86387.1 ABLF02011238 ABLF02015311 ABLF02015320 ABLF02034009 ABLF02043484 LBMM01012412 KMQ86235.1 LBMM01008278 KMQ89009.1 GBXI01013438 JAD00854.1 QOIP01000014 RLU15074.1 GFXV01001748 MBW13553.1 GGMR01006838 MBY19457.1 KU543679 AMS38363.1 KQ972691 KXZ75805.1
AB078931 BAC06456.1 AB078929 BAC06452.1 AB078935 BAC06462.1 LBMM01024882 KMQ82501.1 KZ149896 PZC78733.1 D85594 BAA19776.1 RSAL01000481 RVE41577.1 LBMM01024063 KMQ82623.1 LBMM01012027 KMQ86470.1 KQ460685 KPJ12887.1 LBMM01006063 KMQ90967.1 LBMM01009738 KMQ87908.1 LBMM01019839 KMQ83438.1 LBMM01006954 KMQ90166.1 LBMM01006671 KMQ90407.1 LBMM01005707 KMQ91279.1 LBMM01019838 KMQ83439.1 LBMM01002052 KMQ95336.1 LBMM01004272 KMQ92618.1 LBMM01008448 KMQ88867.1 LBMM01008682 KMQ88705.1 LBMM01006657 KMQ90417.1 LBMM01005733 KMQ91252.1 LBMM01004634 KMQ92224.1 LBMM01021647 KMQ83033.1 LBMM01013668 KMQ85535.1 LBMM01003575 KMQ93366.1 LBMM01007036 KMQ90099.1 LBMM01003302 KMQ93689.1 LBMM01006725 KMQ90373.1 LBMM01004262 KMQ92649.1 LBMM01004261 KMQ92650.1 LBMM01014132 KMQ85305.1 LBMM01007425 KMQ89740.1 GGMR01000779 MBY13398.1 LBMM01003245 KMQ93755.1 LBMM01008422 KMQ88894.1 LBMM01015025 KMQ84953.1 LBMM01007659 KMQ89538.1 QOIP01000010 RLU18098.1 LBMM01006496 KMQ90570.1 QOIP01000031 RLU14697.1 LBMM01003988 KMQ92922.1 LBMM01015018 KMQ84954.1 LBMM01004505 KMQ92346.1 LBMM01004697 KMQ92167.1 LBMM01009527 LBMM01009515 KMQ88059.1 KMQ88067.1 LBMM01004138 KMQ92774.1 LBMM01015285 KMQ84870.1 LBMM01014122 KMQ85315.1 LBMM01005465 KMQ91492.1 GGMR01014195 MBY26814.1 LBMM01002480 KMQ94776.1 LBMM01005216 KMQ91703.1 GBHO01002982 JAG40622.1 LBMM01014956 KMQ84982.1 LBMM01005474 KMQ91485.1 GECU01032129 JAS75577.1 GGMR01015159 MBY27778.1 KMQ94772.1 GGMR01016764 MBY29383.1 LBMM01005845 KMQ91162.1 GGMR01015354 MBY27973.1 ABLF02005203 ABLF02041316 LBMM01012322 LBMM01012138 KMQ86282.1 KMQ86387.1 ABLF02011238 ABLF02015311 ABLF02015320 ABLF02034009 ABLF02043484 LBMM01012412 KMQ86235.1 LBMM01008278 KMQ89009.1 GBXI01013438 JAD00854.1 QOIP01000014 RLU15074.1 GFXV01001748 MBW13553.1 GGMR01006838 MBY19457.1 KU543679 AMS38363.1 KQ972691 KXZ75805.1
Proteomes
Pfam
Interpro
IPR000477
RT_dom
+ More
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR002123 Plipid/glycerol_acylTrfase
IPR012934 Znf_AD
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR029058 AB_hydrolase
IPR013818 Lipase/vitellogenin
IPR000734 TAG_lipase
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR002123 Plipid/glycerol_acylTrfase
IPR012934 Znf_AD
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR029058 AB_hydrolase
IPR013818 Lipase/vitellogenin
IPR000734 TAG_lipase
Gene 3D
ProteinModelPortal
Q868Q4
A0A2W1B333
Q8MY33
Q8MY31
Q8MY35
Q8MY27
+ More
A0A0J7JX58 A0A2W1C3J5 O01419 A0A3S2N998 A0A0J7JWS3 A0A0J7N1D9 A0A194R5D3 A0A0J7KL06 A0A0J7KC11 A0A0J7JZ13 A0A0J7KIY5 A0A0J7KJG6 A0A0J7KM11 A0A0J7JZF7 A0A0J7KYD2 A0A0J7KQK3 A0A0J7KF37 A0A0J7N7S8 A0A0J7KJH4 A0A0J7KLT9 A0A0J7KPR0 A0A0J7JXX4 A0A0J7MYL9 A0A0J7KSR8 A0A0J7KIM6 A0A0J7KTW8 A0A0J7KIW1 A0A0J7KQY4 A0A0J7NJI3 A0A0J7MY07 A0A0J7NAV8 A0A2S2N8I2 A0A0J7NMN5 A0A0J7KF40 A0A0J7K3F1 A0A0J7KGJ6 A0A3L8DDV9 A0A0J7NDI6 A0A3L8D2G3 A0A0J7KRB8 A0A0J7K3R6 A0A0J7KPZ3 A0A0J7KP92 A0A0J7KCZ9 A0A0J7KRB1 A0A0J7MWN5 A0A0J7MY17 A0A0J7KMG5 A0A2S2PBP5 A0A0J7KWU5 A0A0J7KMU1 A0A0A9Z5Y1 A0A0J7K3R2 A0A0J7NFZ0 A0A1B6HLL2 A0A2S2PEZ7 A0A0J7KW81 A0A2S2PIX1 A0A0J7KLK2 A0A2S2PEU2 J9KNG8 A0A0J7K7P2 X1WJY4 J9LJ80 A0A0J7N0R3 A0A0J7KFH2 A0A0A1WPA4 A0A3L8D4C5 A0A2H8THF8 A0A2S2NQQ6 A0A142LX37 A0A139W942
A0A0J7JX58 A0A2W1C3J5 O01419 A0A3S2N998 A0A0J7JWS3 A0A0J7N1D9 A0A194R5D3 A0A0J7KL06 A0A0J7KC11 A0A0J7JZ13 A0A0J7KIY5 A0A0J7KJG6 A0A0J7KM11 A0A0J7JZF7 A0A0J7KYD2 A0A0J7KQK3 A0A0J7KF37 A0A0J7N7S8 A0A0J7KJH4 A0A0J7KLT9 A0A0J7KPR0 A0A0J7JXX4 A0A0J7MYL9 A0A0J7KSR8 A0A0J7KIM6 A0A0J7KTW8 A0A0J7KIW1 A0A0J7KQY4 A0A0J7NJI3 A0A0J7MY07 A0A0J7NAV8 A0A2S2N8I2 A0A0J7NMN5 A0A0J7KF40 A0A0J7K3F1 A0A0J7KGJ6 A0A3L8DDV9 A0A0J7NDI6 A0A3L8D2G3 A0A0J7KRB8 A0A0J7K3R6 A0A0J7KPZ3 A0A0J7KP92 A0A0J7KCZ9 A0A0J7KRB1 A0A0J7MWN5 A0A0J7MY17 A0A0J7KMG5 A0A2S2PBP5 A0A0J7KWU5 A0A0J7KMU1 A0A0A9Z5Y1 A0A0J7K3R2 A0A0J7NFZ0 A0A1B6HLL2 A0A2S2PEZ7 A0A0J7KW81 A0A2S2PIX1 A0A0J7KLK2 A0A2S2PEU2 J9KNG8 A0A0J7K7P2 X1WJY4 J9LJ80 A0A0J7N0R3 A0A0J7KFH2 A0A0A1WPA4 A0A3L8D4C5 A0A2H8THF8 A0A2S2NQQ6 A0A142LX37 A0A139W942
Ontologies
PANTHER
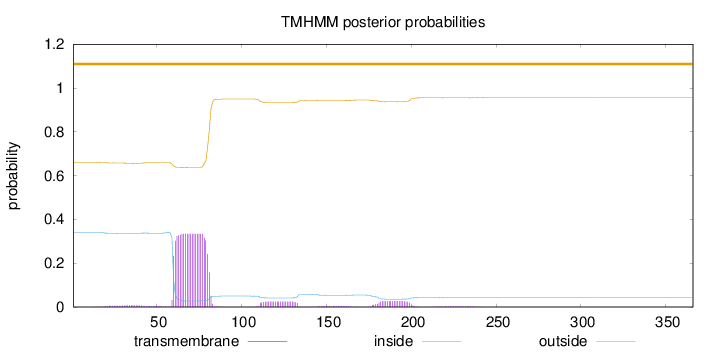
Topology
Subcellular location
Secreted
Length:
366
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.4359
Exp number, first 60 AAs:
0.46801
Total prob of N-in:
0.34038
outside
1 - 366
Population Genetic Test Statistics
Pi
67.599559
Theta
70.636133
Tajima's D
0.61683
CLR
0
CSRT
0.548072596370181
Interpretation
Uncertain
