Gene
KWMTBOMO04302
Pre Gene Modal
BGIBMGA014123
Annotation
PREDICTED:_UDP-xylose_and_UDP-N-acetylglucosamine_transporter_[Papilio_machaon]
Full name
UDP-xylose and UDP-N-acetylglucosamine transporter
Alternative Name
ER GDP-fucose transporter
Solute carrier family 35 member B4 homolog
Solute carrier family 35 member B4 homolog
Location in the cell
PlasmaMembrane Reliability : 4.977
Sequence
CDS
ATGAATTTAAAGGCAGCGCTCGCCATTACTATGGTGTTTGTTGGATGTTGTTCCAACGTAGTTTTTTTGGAGTTTGTCGTCAAAGAGGACCCCGGAGCCGGTAACCTAGCGACCTTCCTGCAGTTTGTATTCATAGCTACGATTGGGTTCTGTACCGTCGGCAAATTCGGTATGGCCAAACGAAATATACCGTTTAGGCAGTATCTACTCCTGGTTGGGTTCTTTTGGACGAGCAGCGTCGCCAACAACTACGCCTTCGACTTTAACATCGCGATGCCGCTGCACATGATCTTCCGAGCTGGTTCTCTAATGGCCAACATGGCCATGGGCGTTTGGATCCTCAAGAAGCGGTACCCACCGCTGAAATACCTGGCCGTGTTTATGATTTCTGCCGGCATAGCCATTTGCACCATTCAGTCTAGCGGAGACGTGAAGGCGCCTCGCAAAACCCACGAGGACGCTGCCGAAGAGGAGAAACTGAAGTTCATCGACTGGCTGTGGTGGTGTTTGGGTATTGCCATACTGACCTTCGCCTTATTCGTCTCGGCTCGGATGGGAATCTTTCAAGAGTCGCTTTATTCGAAGTACGGAAAACACCCGTGGGAGGCATTATACTACACGCATCTTTTGCCATTGATGTTCTGGTTGCCGACCGCTTCGAATCTGATCGGACACGTCGAAATCGCGACAGAAACCCCTGCCGTCGAGTTCTTAGGGTTCATTTTGCCACGACAAGTTCTGTGGCTGTTGCTGTACGTTTCGACTCAGGGTCTGTGTATTAGCGCGGTCTACGTTTTGACAACCGAATGTGCGTCGCTGACAGTGACGTTAGCTGTCACTTTGAGAAAGTTCGTGTCGCTTCTGTTCTCTATAATGTACTTCAGGAATCCCTTCACTTTAGGGCACTGGATAGGGACATTGCTAGTGTTCGTCGGTACGTTGATATTCACGGAGATATTGCAGAAATGCTTGGCCGTGTTTACTACAGACGATAAGAAGAAAAAATAA
Protein
MNLKAALAITMVFVGCCSNVVFLEFVVKEDPGAGNLATFLQFVFIATIGFCTVGKFGMAKRNIPFRQYLLLVGFFWTSSVANNYAFDFNIAMPLHMIFRAGSLMANMAMGVWILKKRYPPLKYLAVFMISAGIAICTIQSSGDVKAPRKTHEDAAEEEKLKFIDWLWWCLGIAILTFALFVSARMGIFQESLYSKYGKHPWEALYYTHLLPLMFWLPTASNLIGHVEIATETPAVEFLGFILPRQVLWLLLYVSTQGLCISAVYVLTTECASLTVTLAVTLRKFVSLLFSIMYFRNPFTLGHWIGTLLVFVGTLIFTEILQKCLAVFTTDDKKKK
Summary
Description
Sugar transporter that specifically mediates the transport of UDP-N-acetylglucosamine (UDP-GlcNAc), GDP-fucose and UDP-xylose. Functions redundantly with nac in the O-fucosylation of Notch, positively regulating Notch signaling. Involved in the biosynthesis of heparan sulfate-glycosaminoglycan (HS-GAG) and in Dpp signaling in the wing imaginal disk.
Similarity
Belongs to the nucleotide-sugar transporter family. SLC35B subfamily.
Keywords
Complete proteome
Endoplasmic reticulum
Membrane
Reference proteome
Sugar transport
Transmembrane
Transmembrane helix
Transport
Feature
chain UDP-xylose and UDP-N-acetylglucosamine transporter
Uniprot
H9JX58
A0A194RFF5
A0A194Q204
A0A2H1VJI7
A0A1Z2RRJ7
A0A1E1W6N0
+ More
S4NNN6 A0A212EP67 A0A0L7L6J9 A0A0K8TQS5 A0A182XC69 A0A182LPE1 A0A2C9GQT5 Q7Q357 A0A182LTW9 A0A182VGY5 A0A336K9S9 A0A2M4BT84 B4NPD8 A0A2M3Z9R0 A0A2M3ZE64 T1DNH9 A0A2M4BT00 Q174K4 W5JJP8 A0A182QE09 A0A2M4AQ20 A0A084WB95 J9KAW4 B0WBU7 A0A2J7QCY3 A0A067R3Y0 A0A182JXG0 A0A154PF24 A0A182J086 C4WUT8 A0A1Q3FHN4 A0A182PVC9 B3MQ47 A0A1Q3FHT1 A0A1B0CTY7 A0A0L7QU21 E1ZV25 A0A1Q3FHF1 A0A182FU17 B4M864 B4L6J2 E2B348 A0A182YJZ8 A4V408 Q9W429 B4MH52 A0A3B0JW76 B4R5D8 A0A0R1E9Q2 B4PYP7 B3NXP4 B4HB97 Q29GG2 A0A182MY20 A0A026X0H2 A0A0C9Q2C5 B4L6J3 A0A3L8DS66 A0A0K8U8G2 B4JNK7 A0A1B6MFC4 T1P969 A0A1L8DEL9 A0A1B6C7M0 A0A0T6BCG0 A0A195AVF8 A0A158P260 A0A151IQU3 A0A1W4VSE8 W8CA71 A0A1I8MZC4 A0A0M9A234 A0A1L8DEW4 A0A195FJ21 A0A2A3E4K2 A0A0L0CKV5 A0A195DMA6 U5EU57 B4MBW8 A0A1I8PE78 A0A1I8PEG5 B4JNK8 A0A3B3RQX3 E9FSD0 D7EIK0 K7IR88 A0A0M4EUI7 A0A315VYV1 I3JPR9 A0A3Q3BJK3 A0A3B5ATC2 A0A3B3VZD6
S4NNN6 A0A212EP67 A0A0L7L6J9 A0A0K8TQS5 A0A182XC69 A0A182LPE1 A0A2C9GQT5 Q7Q357 A0A182LTW9 A0A182VGY5 A0A336K9S9 A0A2M4BT84 B4NPD8 A0A2M3Z9R0 A0A2M3ZE64 T1DNH9 A0A2M4BT00 Q174K4 W5JJP8 A0A182QE09 A0A2M4AQ20 A0A084WB95 J9KAW4 B0WBU7 A0A2J7QCY3 A0A067R3Y0 A0A182JXG0 A0A154PF24 A0A182J086 C4WUT8 A0A1Q3FHN4 A0A182PVC9 B3MQ47 A0A1Q3FHT1 A0A1B0CTY7 A0A0L7QU21 E1ZV25 A0A1Q3FHF1 A0A182FU17 B4M864 B4L6J2 E2B348 A0A182YJZ8 A4V408 Q9W429 B4MH52 A0A3B0JW76 B4R5D8 A0A0R1E9Q2 B4PYP7 B3NXP4 B4HB97 Q29GG2 A0A182MY20 A0A026X0H2 A0A0C9Q2C5 B4L6J3 A0A3L8DS66 A0A0K8U8G2 B4JNK7 A0A1B6MFC4 T1P969 A0A1L8DEL9 A0A1B6C7M0 A0A0T6BCG0 A0A195AVF8 A0A158P260 A0A151IQU3 A0A1W4VSE8 W8CA71 A0A1I8MZC4 A0A0M9A234 A0A1L8DEW4 A0A195FJ21 A0A2A3E4K2 A0A0L0CKV5 A0A195DMA6 U5EU57 B4MBW8 A0A1I8PE78 A0A1I8PEG5 B4JNK8 A0A3B3RQX3 E9FSD0 D7EIK0 K7IR88 A0A0M4EUI7 A0A315VYV1 I3JPR9 A0A3Q3BJK3 A0A3B5ATC2 A0A3B3VZD6
Pubmed
19121390
26354079
28605547
23622113
22118469
26227816
+ More
26369729 20966253 12364791 14747013 17210077 17994087 17510324 20920257 23761445 24438588 24845553 18057021 20798317 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19948734 17550304 15632085 23185243 24508170 30249741 21347285 24495485 25315136 26108605 29240929 21292972 18362917 19820115 20075255 29703783 25186727
26369729 20966253 12364791 14747013 17210077 17994087 17510324 20920257 23761445 24438588 24845553 18057021 20798317 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19948734 17550304 15632085 23185243 24508170 30249741 21347285 24495485 25315136 26108605 29240929 21292972 18362917 19820115 20075255 29703783 25186727
EMBL
BABH01031954
KQ460211
KPJ16558.1
KQ459580
KPI99363.1
ODYU01002910
+ More
SOQ40995.1 KY938828 ASA46469.1 GDQN01008416 JAT82638.1 GAIX01013856 JAA78704.1 AGBW02013518 OWR43274.1 JTDY01002683 KOB70936.1 GDAI01000894 JAI16709.1 APCN01001930 AAAB01008966 EAA13009.4 AXCM01000360 UFQS01000100 UFQT01000100 SSW99523.1 SSX19903.1 GGFJ01007073 MBW56214.1 CH964291 EDW86378.1 GGFM01004525 MBW25276.1 GGFM01006085 MBW26836.1 GAMD01002860 JAA98730.1 GGFJ01007075 MBW56216.1 CH477409 EAT41500.1 EAT41501.1 ADMH02001328 ETN62994.1 AXCN02001640 GGFK01009538 MBW42859.1 ATLV01022327 KE525331 KFB47489.1 ABLF02031956 DS231882 EDS42877.1 NEVH01015855 PNF26435.1 KK852715 KDR17872.1 KQ434885 KZC10044.1 AK341218 BAH71658.1 GFDL01007978 JAV27067.1 CH902621 EDV44473.1 KPU81612.1 GFDL01007919 JAV27126.1 AJWK01028174 KQ414740 KOC62054.1 GL434395 EFN74954.1 GFDL01008112 JAV26933.1 CH940653 EDW62340.1 CH933812 EDW05988.1 KRG07109.1 GL445285 EFN89897.1 AE014298 AAN09162.1 BT021217 CH941246 EDW71473.1 OUUW01000011 SPP86335.1 CM000366 EDX17234.1 CM000162 KRK05861.1 KRK05862.1 KRK05863.1 EDX00983.2 CH954180 EDV47345.1 CH479248 EDW37909.1 CH379064 EAL32147.1 KRT06996.1 KRT06997.1 KK107054 EZA61573.1 GBYB01008153 JAG77920.1 EDW05989.1 QOIP01000004 RLU23285.1 GDHF01029446 GDHF01016511 JAI22868.1 JAI35803.1 CH916371 EDV92300.1 GEBQ01005385 JAT34592.1 KA645249 AFP59878.1 GFDF01009193 JAV04891.1 GEDC01027884 JAS09414.1 LJIG01002143 KRT84797.1 KQ976731 KYM76218.1 ADTU01006992 KQ976755 KYN08577.1 GAMC01001541 GAMC01001540 GAMC01001539 JAC05017.1 KQ435778 KOX74846.1 GFDF01009194 JAV04890.1 KQ981523 KYN40366.1 KZ288387 PBC26414.1 JRES01000340 KNC32109.1 KQ980724 KYN14035.1 GANO01002473 JAB57398.1 CH940656 EDW58589.1 EDV92301.1 GL732524 EFX89856.1 KQ971307 EFA12028.1 CP012528 ALC48760.1 NHOQ01000831 PWA28500.1 AERX01010942
SOQ40995.1 KY938828 ASA46469.1 GDQN01008416 JAT82638.1 GAIX01013856 JAA78704.1 AGBW02013518 OWR43274.1 JTDY01002683 KOB70936.1 GDAI01000894 JAI16709.1 APCN01001930 AAAB01008966 EAA13009.4 AXCM01000360 UFQS01000100 UFQT01000100 SSW99523.1 SSX19903.1 GGFJ01007073 MBW56214.1 CH964291 EDW86378.1 GGFM01004525 MBW25276.1 GGFM01006085 MBW26836.1 GAMD01002860 JAA98730.1 GGFJ01007075 MBW56216.1 CH477409 EAT41500.1 EAT41501.1 ADMH02001328 ETN62994.1 AXCN02001640 GGFK01009538 MBW42859.1 ATLV01022327 KE525331 KFB47489.1 ABLF02031956 DS231882 EDS42877.1 NEVH01015855 PNF26435.1 KK852715 KDR17872.1 KQ434885 KZC10044.1 AK341218 BAH71658.1 GFDL01007978 JAV27067.1 CH902621 EDV44473.1 KPU81612.1 GFDL01007919 JAV27126.1 AJWK01028174 KQ414740 KOC62054.1 GL434395 EFN74954.1 GFDL01008112 JAV26933.1 CH940653 EDW62340.1 CH933812 EDW05988.1 KRG07109.1 GL445285 EFN89897.1 AE014298 AAN09162.1 BT021217 CH941246 EDW71473.1 OUUW01000011 SPP86335.1 CM000366 EDX17234.1 CM000162 KRK05861.1 KRK05862.1 KRK05863.1 EDX00983.2 CH954180 EDV47345.1 CH479248 EDW37909.1 CH379064 EAL32147.1 KRT06996.1 KRT06997.1 KK107054 EZA61573.1 GBYB01008153 JAG77920.1 EDW05989.1 QOIP01000004 RLU23285.1 GDHF01029446 GDHF01016511 JAI22868.1 JAI35803.1 CH916371 EDV92300.1 GEBQ01005385 JAT34592.1 KA645249 AFP59878.1 GFDF01009193 JAV04891.1 GEDC01027884 JAS09414.1 LJIG01002143 KRT84797.1 KQ976731 KYM76218.1 ADTU01006992 KQ976755 KYN08577.1 GAMC01001541 GAMC01001540 GAMC01001539 JAC05017.1 KQ435778 KOX74846.1 GFDF01009194 JAV04890.1 KQ981523 KYN40366.1 KZ288387 PBC26414.1 JRES01000340 KNC32109.1 KQ980724 KYN14035.1 GANO01002473 JAB57398.1 CH940656 EDW58589.1 EDV92301.1 GL732524 EFX89856.1 KQ971307 EFA12028.1 CP012528 ALC48760.1 NHOQ01000831 PWA28500.1 AERX01010942
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000037510
UP000076407
+ More
UP000075882 UP000075840 UP000007062 UP000075883 UP000075903 UP000007798 UP000008820 UP000000673 UP000075886 UP000030765 UP000007819 UP000002320 UP000235965 UP000027135 UP000075881 UP000076502 UP000075880 UP000075885 UP000007801 UP000092461 UP000053825 UP000000311 UP000069272 UP000008792 UP000009192 UP000008237 UP000076408 UP000000803 UP000268350 UP000000304 UP000002282 UP000008711 UP000008744 UP000001819 UP000075884 UP000053097 UP000279307 UP000001070 UP000078540 UP000005205 UP000078542 UP000192221 UP000095301 UP000053105 UP000078541 UP000242457 UP000037069 UP000078492 UP000095300 UP000261540 UP000000305 UP000007266 UP000002358 UP000092553 UP000005207 UP000264840 UP000261400 UP000261500
UP000075882 UP000075840 UP000007062 UP000075883 UP000075903 UP000007798 UP000008820 UP000000673 UP000075886 UP000030765 UP000007819 UP000002320 UP000235965 UP000027135 UP000075881 UP000076502 UP000075880 UP000075885 UP000007801 UP000092461 UP000053825 UP000000311 UP000069272 UP000008792 UP000009192 UP000008237 UP000076408 UP000000803 UP000268350 UP000000304 UP000002282 UP000008711 UP000008744 UP000001819 UP000075884 UP000053097 UP000279307 UP000001070 UP000078540 UP000005205 UP000078542 UP000192221 UP000095301 UP000053105 UP000078541 UP000242457 UP000037069 UP000078492 UP000095300 UP000261540 UP000000305 UP000007266 UP000002358 UP000092553 UP000005207 UP000264840 UP000261400 UP000261500
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JX58
A0A194RFF5
A0A194Q204
A0A2H1VJI7
A0A1Z2RRJ7
A0A1E1W6N0
+ More
S4NNN6 A0A212EP67 A0A0L7L6J9 A0A0K8TQS5 A0A182XC69 A0A182LPE1 A0A2C9GQT5 Q7Q357 A0A182LTW9 A0A182VGY5 A0A336K9S9 A0A2M4BT84 B4NPD8 A0A2M3Z9R0 A0A2M3ZE64 T1DNH9 A0A2M4BT00 Q174K4 W5JJP8 A0A182QE09 A0A2M4AQ20 A0A084WB95 J9KAW4 B0WBU7 A0A2J7QCY3 A0A067R3Y0 A0A182JXG0 A0A154PF24 A0A182J086 C4WUT8 A0A1Q3FHN4 A0A182PVC9 B3MQ47 A0A1Q3FHT1 A0A1B0CTY7 A0A0L7QU21 E1ZV25 A0A1Q3FHF1 A0A182FU17 B4M864 B4L6J2 E2B348 A0A182YJZ8 A4V408 Q9W429 B4MH52 A0A3B0JW76 B4R5D8 A0A0R1E9Q2 B4PYP7 B3NXP4 B4HB97 Q29GG2 A0A182MY20 A0A026X0H2 A0A0C9Q2C5 B4L6J3 A0A3L8DS66 A0A0K8U8G2 B4JNK7 A0A1B6MFC4 T1P969 A0A1L8DEL9 A0A1B6C7M0 A0A0T6BCG0 A0A195AVF8 A0A158P260 A0A151IQU3 A0A1W4VSE8 W8CA71 A0A1I8MZC4 A0A0M9A234 A0A1L8DEW4 A0A195FJ21 A0A2A3E4K2 A0A0L0CKV5 A0A195DMA6 U5EU57 B4MBW8 A0A1I8PE78 A0A1I8PEG5 B4JNK8 A0A3B3RQX3 E9FSD0 D7EIK0 K7IR88 A0A0M4EUI7 A0A315VYV1 I3JPR9 A0A3Q3BJK3 A0A3B5ATC2 A0A3B3VZD6
S4NNN6 A0A212EP67 A0A0L7L6J9 A0A0K8TQS5 A0A182XC69 A0A182LPE1 A0A2C9GQT5 Q7Q357 A0A182LTW9 A0A182VGY5 A0A336K9S9 A0A2M4BT84 B4NPD8 A0A2M3Z9R0 A0A2M3ZE64 T1DNH9 A0A2M4BT00 Q174K4 W5JJP8 A0A182QE09 A0A2M4AQ20 A0A084WB95 J9KAW4 B0WBU7 A0A2J7QCY3 A0A067R3Y0 A0A182JXG0 A0A154PF24 A0A182J086 C4WUT8 A0A1Q3FHN4 A0A182PVC9 B3MQ47 A0A1Q3FHT1 A0A1B0CTY7 A0A0L7QU21 E1ZV25 A0A1Q3FHF1 A0A182FU17 B4M864 B4L6J2 E2B348 A0A182YJZ8 A4V408 Q9W429 B4MH52 A0A3B0JW76 B4R5D8 A0A0R1E9Q2 B4PYP7 B3NXP4 B4HB97 Q29GG2 A0A182MY20 A0A026X0H2 A0A0C9Q2C5 B4L6J3 A0A3L8DS66 A0A0K8U8G2 B4JNK7 A0A1B6MFC4 T1P969 A0A1L8DEL9 A0A1B6C7M0 A0A0T6BCG0 A0A195AVF8 A0A158P260 A0A151IQU3 A0A1W4VSE8 W8CA71 A0A1I8MZC4 A0A0M9A234 A0A1L8DEW4 A0A195FJ21 A0A2A3E4K2 A0A0L0CKV5 A0A195DMA6 U5EU57 B4MBW8 A0A1I8PE78 A0A1I8PEG5 B4JNK8 A0A3B3RQX3 E9FSD0 D7EIK0 K7IR88 A0A0M4EUI7 A0A315VYV1 I3JPR9 A0A3Q3BJK3 A0A3B5ATC2 A0A3B3VZD6
Ontologies
GO
PANTHER
Topology
Subcellular location
Endoplasmic reticulum membrane
SignalP
Position: 1 - 18,
Likelihood: 0.543140
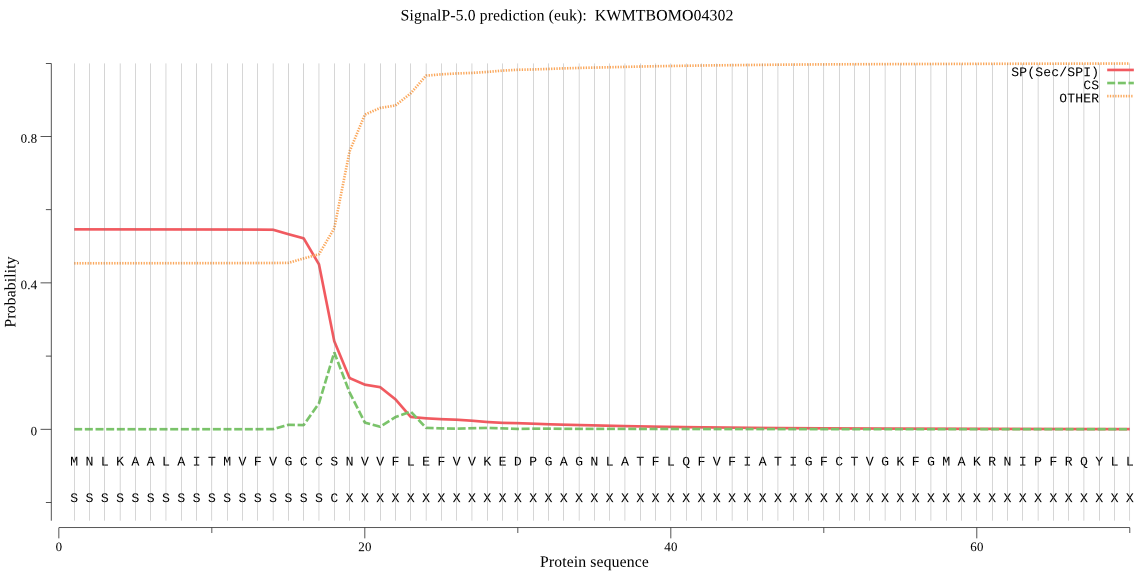
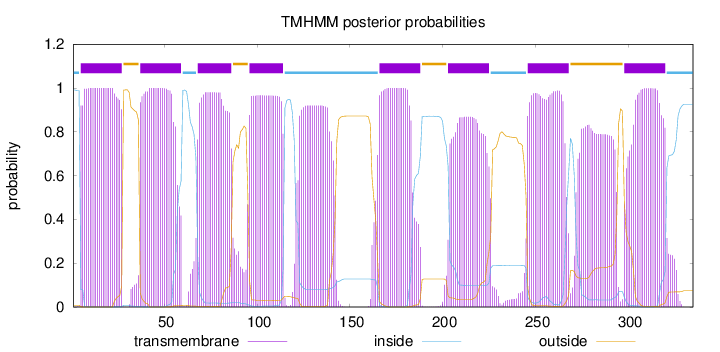
Length:
335
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
204.58497
Exp number, first 60 AAs:
43.99553
Total prob of N-in:
0.99173
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 36
TMhelix
37 - 59
inside
60 - 67
TMhelix
68 - 86
outside
87 - 95
TMhelix
96 - 114
inside
115 - 165
TMhelix
166 - 188
outside
189 - 202
TMhelix
203 - 225
inside
226 - 245
TMhelix
246 - 268
outside
269 - 297
TMhelix
298 - 320
inside
321 - 335
Population Genetic Test Statistics
Pi
230.963706
Theta
213.205857
Tajima's D
0.23076
CLR
0
CSRT
0.428328583570821
Interpretation
Uncertain
