Gene
KWMTBOMO04300 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014116
Annotation
beta-N-acetylglucosaminidase_3_precursor_[Bombyx_mori]
Full name
Beta-hexosaminidase
Location in the cell
Cytoplasmic Reliability : 2.041
Sequence
CDS
ATGGAAATAAGCGATATTTTAGGCGCTTTTTTAGTGACCGGTTTGCATATCGTGGAACCGGGACCCGAATATCCAGCATCGAAAGGTGCAATTTGGCCAAGACCGCAAATGCAATCGATAGAAATTCCTTATTACAAATTTGATAGTGATATACTCGAAATTAAGGTAGTCGATCACGATTGCCCTATTCTGTCGAATGCCGTCCAACGGAGTCTGGCGGTACTGAGAGAGATGCTCCGAATCGCGAGTCCGTATGTAAACCGCAATGCGCCACAACAGGTCTTAGATGATGATACATACGACGGACCACTGAAGAGTCTAAGCATTTACTTGACATCACCCTGTGAAGAGTATCCGCATTTCGGCATGATCGAGAGCTACAATCTGACGATCGCCGCTGACAGCACACTCAGGAGCTCATCGATATGGGGAATACTGAGAGGCTTGGAAAGTTGGACGCACCTGTTTCATCTCTCAGATAATCGTGATCAATTGCACATTAACAAAGGTGAAGTTCACGACTTTCCCCGTTACGCTCACAGAGGACTGCTCGTTGACACATCACGTCATTACATCTCAATGTCTAATATCCTACTCATTTTGGACGCTATGGCCATGAACAAGATGAACGTTTTTCACTGGCACATTGTTGACGACCAGAGCTTCCCATATCAGAGCGAAAGGTTTCCGGATTTAAGACAGTCTATAAGAATCGTTGGTCTAGGTCACACAAGGTCCTGGGGTGTAGCCAAACCCGACCTCCTGACACATTGCTACGACCAAGACGGTGACTACGTGGGTCTCGGTCCGATGAACCCAATCAAGGACAGCACATACACCTTCCTGCAGGAACTGTTTCATGAGGTCCAGGCCTTGTTTCCGGATAGATACATTCACATCGGAGGTGATGAAGTCGATCTAGATTGCTGGGAGTCGAATCCAGAATTCCAGAGATACATACAAGAGCACAACCTGACTTCGGTAGCTGATTTTCACGCTCTATTCATGCGCAACACGATTCCTCTGCTCAGCGAAAACTCGAGACCGATAGTGTGGCAGGAAGTTTTCGATGAAGGCGTGCCCCTTCCGAAGGACACTATCGTGCAAGTGTGGAAGGAAAACGAAGCTCCTGAAATGTTGAATATCCTAAGAGCAAGTCACCAGTTAATCTATTCCACTGGCTGGTACCTTGATCATCTGAACACTGGAGGCGACTGGACGGAATTTTTCAACAAGGACCCTCGGGACCTGGTCAACGGCCTCAGCAAAGACATCAATGTTGACAACATTGTAGGTGGCGAAGCTTGCATGTGGGCAGAAGTTGTTAACGACATGAACATTATGAGCAGAGTATGGCCACGAGCTAGTGCTGTAGCTGAAAGACTGTGGGGCCACGAATCACAGGCAACATATCAAGTACACTGTCGCTTAGAGGAGCACACGTGCAGAATGAACGCGCGCGGCATACATGCCCAGCCACCCAGCGGCCCTGGCTTCTGCCTAGGCGTTTAG
Protein
MEISDILGAFLVTGLHIVEPGPEYPASKGAIWPRPQMQSIEIPYYKFDSDILEIKVVDHDCPILSNAVQRSLAVLREMLRIASPYVNRNAPQQVLDDDTYDGPLKSLSIYLTSPCEEYPHFGMIESYNLTIAADSTLRSSSIWGILRGLESWTHLFHLSDNRDQLHINKGEVHDFPRYAHRGLLVDTSRHYISMSNILLILDAMAMNKMNVFHWHIVDDQSFPYQSERFPDLRQSIRIVGLGHTRSWGVAKPDLLTHCYDQDGDYVGLGPMNPIKDSTYTFLQELFHEVQALFPDRYIHIGGDEVDLDCWESNPEFQRYIQEHNLTSVADFHALFMRNTIPLLSENSRPIVWQEVFDEGVPLPKDTIVQVWKENEAPEMLNILRASHQLIYSTGWYLDHLNTGGDWTEFFNKDPRDLVNGLSKDINVDNIVGGEACMWAEVVNDMNIMSRVWPRASAVAERLWGHESQATYQVHCRLEEHTCRMNARGIHAQPPSGPGFCLGV
Summary
Catalytic Activity
Hydrolysis of terminal non-reducing N-acetyl-D-hexosamine residues in N-acetyl-beta-D-hexosaminides.
Similarity
Belongs to the glycosyl hydrolase 20 family.
Feature
chain Beta-hexosaminidase
Uniprot
Q3L6N4
H9JX51
A4PHN7
Q3L6N3
H9JX50
A4LAF9
+ More
Q2Q1D3 Q3LS76 Q2Q1D2 Q3LS75 A0A194QY72 A0A212F3V9 A0A194QY01 A0A194PM72 A0A2H1V9E6 B6CI24 B0XBN5 J9HI66 Q17BL1 A0A182KFH4 A0A182QVJ5 A0A182VFS0 A0A182HR23 A0A182W8J9 A0A182L7V3 A0A1L8DQZ7 A0A0L7KE68 B0XBN6 A0A182Y637 A0A182JLR9 A0A026WT31 E0VQW8 Q7Q0Z2 U5ETX4 A0A3G5BIJ3 W5JJY2 A0A1W6EW10 A0A1B0CZC5 A0A182R1R5 D6WRR7 A0A1Y1LY98 A0A1J1HQF6 A0A087ZUU5 A0A084W3Y6 A0A0M8ZZH8 A0A2J7PCV4 A0A0L7QS85 A0A1B6CMM3 A0A2P8Y3J8 D6WRR9 A0A182PTQ9 A0A182UFT6 A0A182XHA2 D6WRR8 A0A182NNT6 A0A182GID0 U4TU30 N6T5A7 A0A2J7PCU7 A0A182FN54 A0A067R5X1 A0A1W4XBS7 V5H217 A0A182LTR5 A0A147B289 A0A1B6KR81 A0A146Z1P6 A0A3Q2QBK9 A0A1B6KBG0 A0A0C9RX26 A0A1S3HJI4 B7PE53 A0A3Q1IDG1 A0A1D2MNV3 A0A0K2SW92 A0A131YVJ8 A0A1S3HGE0 A0A1B6EVU7 A0A3Q1EKY7 A0A1S3HI83 V3ZMB9 Q7YTB2 L7MG67 A0A224YPI0 A0A3B1J6J9 A0A3P8XAC0 A0A131XKM5 A0A1S3HGG6 A0A3Q3G324 A0A3B3Q349 A0A2D0QB43 A0A3M7R7F3 A0A3B1J888 A0A3Q1CKD5 A0A3Q1H4L2 A0A2R5LAG8 A0A0A1WSN4 A0A131XIX2 A0A3Q1I5U8 A0A3B1K7J4
Q2Q1D3 Q3LS76 Q2Q1D2 Q3LS75 A0A194QY72 A0A212F3V9 A0A194QY01 A0A194PM72 A0A2H1V9E6 B6CI24 B0XBN5 J9HI66 Q17BL1 A0A182KFH4 A0A182QVJ5 A0A182VFS0 A0A182HR23 A0A182W8J9 A0A182L7V3 A0A1L8DQZ7 A0A0L7KE68 B0XBN6 A0A182Y637 A0A182JLR9 A0A026WT31 E0VQW8 Q7Q0Z2 U5ETX4 A0A3G5BIJ3 W5JJY2 A0A1W6EW10 A0A1B0CZC5 A0A182R1R5 D6WRR7 A0A1Y1LY98 A0A1J1HQF6 A0A087ZUU5 A0A084W3Y6 A0A0M8ZZH8 A0A2J7PCV4 A0A0L7QS85 A0A1B6CMM3 A0A2P8Y3J8 D6WRR9 A0A182PTQ9 A0A182UFT6 A0A182XHA2 D6WRR8 A0A182NNT6 A0A182GID0 U4TU30 N6T5A7 A0A2J7PCU7 A0A182FN54 A0A067R5X1 A0A1W4XBS7 V5H217 A0A182LTR5 A0A147B289 A0A1B6KR81 A0A146Z1P6 A0A3Q2QBK9 A0A1B6KBG0 A0A0C9RX26 A0A1S3HJI4 B7PE53 A0A3Q1IDG1 A0A1D2MNV3 A0A0K2SW92 A0A131YVJ8 A0A1S3HGE0 A0A1B6EVU7 A0A3Q1EKY7 A0A1S3HI83 V3ZMB9 Q7YTB2 L7MG67 A0A224YPI0 A0A3B1J6J9 A0A3P8XAC0 A0A131XKM5 A0A1S3HGG6 A0A3Q3G324 A0A3B3Q349 A0A2D0QB43 A0A3M7R7F3 A0A3B1J888 A0A3Q1CKD5 A0A3Q1H4L2 A0A2R5LAG8 A0A0A1WSN4 A0A131XIX2 A0A3Q1I5U8 A0A3B1K7J4
EC Number
3.2.1.52
Pubmed
19121390
17617724
16427309
16684772
26354079
22118469
+ More
17510324 20966253 26227816 25244985 24508170 30249741 20566863 12364791 30400621 20920257 23761445 18362917 19820115 28004739 24438588 29403074 26483478 23537049 24845553 25765539 27289101 26830274 23254933 12828682 25576852 28797301 25329095 25069045 28049606 29240929 30375419 25830018
17510324 20966253 26227816 25244985 24508170 30249741 20566863 12364791 30400621 20920257 23761445 18362917 19820115 28004739 24438588 29403074 26483478 23537049 24845553 25765539 27289101 26830274 23254933 12828682 25576852 28797301 25329095 25069045 28049606 29240929 30375419 25830018
EMBL
AY601817
AAT99455.1
BABH01031930
BABH01031931
BABH01031932
AB286958
+ More
BAF52532.1 AY601818 AAT99456.1 BABH01031937 BABH01031938 EF469203 ABO65045.1 DQ249307 ABB76924.1 DQ183186 DQ249309 ABA27426.1 ABB76926.1 DQ249308 ABB76925.1 DQ183187 ABA27427.1 KQ460949 KPJ10417.1 AGBW02010485 OWR48417.1 KPJ10418.1 KQ459598 KPI94536.1 ODYU01001373 SOQ37455.1 EU180214 ABY57947.1 DS232645 EDS44384.1 CH477320 EJY57559.1 EAT43655.1 AXCN02001981 APCN01001633 GFDF01005290 JAV08794.1 JTDY01010100 KOB61159.1 EDS44385.1 KK107128 QOIP01000006 EZA58264.1 RLU21577.1 DS235442 EEB15774.1 AAAB01008980 EAA14547.4 GANO01002584 JAB57287.1 MK075207 AYV99610.1 ADMH02001300 ETN63089.1 KY563514 ARK19923.1 AJVK01009523 AJVK01009524 KQ971354 EFA05960.2 GEZM01046313 JAV77340.1 CVRI01000008 CRK88449.1 ATLV01020229 ATLV01020230 KE525296 KFB44930.1 KQ435792 KOX74285.1 NEVH01026403 PNF14160.1 KQ414768 KOC61341.1 GEDC01022680 JAS14618.1 PYGN01000972 PSN38811.1 EFA07069.1 EFA05959.1 JXUM01066149 KQ562383 KXJ76018.1 KB631654 ERL85064.1 APGK01052022 APGK01052023 KB741208 ENN72853.1 PNF14159.1 KK852722 KDR17734.1 GANP01013509 JAB70959.1 AXCM01006623 GCES01001496 JAR84827.1 GEBQ01026015 JAT13962.1 GCES01026591 JAR59732.1 GEBQ01031184 JAT08793.1 GBYB01012436 JAG82203.1 ABJB010025437 ABJB010031690 ABJB010274095 ABJB010318523 ABJB010440044 ABJB010990743 ABJB011081853 ABJB011094047 ABJB011106130 ABJB011126424 DS693693 EEC04875.1 LJIJ01000794 ODM94535.1 HACA01000653 CDW18014.1 GEDV01005318 JAP83239.1 GECZ01027707 JAS42062.1 KB202050 ESO92513.1 AJ518021 CAD57204.1 GACK01002776 JAA62258.1 GFPF01008361 MAA19507.1 GEFH01001454 JAP67127.1 REGN01004092 RNA19155.1 GGLE01002319 MBY06445.1 GBXI01012203 JAD02089.1 GEFH01002269 JAP66312.1
BAF52532.1 AY601818 AAT99456.1 BABH01031937 BABH01031938 EF469203 ABO65045.1 DQ249307 ABB76924.1 DQ183186 DQ249309 ABA27426.1 ABB76926.1 DQ249308 ABB76925.1 DQ183187 ABA27427.1 KQ460949 KPJ10417.1 AGBW02010485 OWR48417.1 KPJ10418.1 KQ459598 KPI94536.1 ODYU01001373 SOQ37455.1 EU180214 ABY57947.1 DS232645 EDS44384.1 CH477320 EJY57559.1 EAT43655.1 AXCN02001981 APCN01001633 GFDF01005290 JAV08794.1 JTDY01010100 KOB61159.1 EDS44385.1 KK107128 QOIP01000006 EZA58264.1 RLU21577.1 DS235442 EEB15774.1 AAAB01008980 EAA14547.4 GANO01002584 JAB57287.1 MK075207 AYV99610.1 ADMH02001300 ETN63089.1 KY563514 ARK19923.1 AJVK01009523 AJVK01009524 KQ971354 EFA05960.2 GEZM01046313 JAV77340.1 CVRI01000008 CRK88449.1 ATLV01020229 ATLV01020230 KE525296 KFB44930.1 KQ435792 KOX74285.1 NEVH01026403 PNF14160.1 KQ414768 KOC61341.1 GEDC01022680 JAS14618.1 PYGN01000972 PSN38811.1 EFA07069.1 EFA05959.1 JXUM01066149 KQ562383 KXJ76018.1 KB631654 ERL85064.1 APGK01052022 APGK01052023 KB741208 ENN72853.1 PNF14159.1 KK852722 KDR17734.1 GANP01013509 JAB70959.1 AXCM01006623 GCES01001496 JAR84827.1 GEBQ01026015 JAT13962.1 GCES01026591 JAR59732.1 GEBQ01031184 JAT08793.1 GBYB01012436 JAG82203.1 ABJB010025437 ABJB010031690 ABJB010274095 ABJB010318523 ABJB010440044 ABJB010990743 ABJB011081853 ABJB011094047 ABJB011106130 ABJB011126424 DS693693 EEC04875.1 LJIJ01000794 ODM94535.1 HACA01000653 CDW18014.1 GEDV01005318 JAP83239.1 GECZ01027707 JAS42062.1 KB202050 ESO92513.1 AJ518021 CAD57204.1 GACK01002776 JAA62258.1 GFPF01008361 MAA19507.1 GEFH01001454 JAP67127.1 REGN01004092 RNA19155.1 GGLE01002319 MBY06445.1 GBXI01012203 JAD02089.1 GEFH01002269 JAP66312.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000053268
UP000002320
UP000008820
+ More
UP000075881 UP000075886 UP000075903 UP000075840 UP000075920 UP000075882 UP000037510 UP000076408 UP000075880 UP000053097 UP000279307 UP000009046 UP000007062 UP000000673 UP000092462 UP000075900 UP000007266 UP000183832 UP000005203 UP000030765 UP000053105 UP000235965 UP000053825 UP000245037 UP000075885 UP000075902 UP000076407 UP000075884 UP000069940 UP000249989 UP000030742 UP000019118 UP000069272 UP000027135 UP000192223 UP000075883 UP000265000 UP000085678 UP000001555 UP000265040 UP000094527 UP000257200 UP000030746 UP000018467 UP000265140 UP000261660 UP000261540 UP000221080 UP000276133 UP000257160
UP000075881 UP000075886 UP000075903 UP000075840 UP000075920 UP000075882 UP000037510 UP000076408 UP000075880 UP000053097 UP000279307 UP000009046 UP000007062 UP000000673 UP000092462 UP000075900 UP000007266 UP000183832 UP000005203 UP000030765 UP000053105 UP000235965 UP000053825 UP000245037 UP000075885 UP000075902 UP000076407 UP000075884 UP000069940 UP000249989 UP000030742 UP000019118 UP000069272 UP000027135 UP000192223 UP000075883 UP000265000 UP000085678 UP000001555 UP000265040 UP000094527 UP000257200 UP000030746 UP000018467 UP000265140 UP000261660 UP000261540 UP000221080 UP000276133 UP000257160
Pfam
Interpro
IPR017853
Glycoside_hydrolase_SF
+ More
IPR025705 Beta_hexosaminidase_sua/sub
IPR029018 Hex-like_dom2
IPR015883 Glyco_hydro_20_cat
IPR029019 HEX_eukaryotic_N
IPR036412 HAD-like_sf
IPR004274 FCP1_dom
IPR036734 Neur_chan_lig-bd_sf
IPR006202 Neur_chan_lig-bd
IPR004625 PyrdxlKinase
IPR013749 PM/HMP-P_kinase-1
IPR029056 Ribokinase-like
IPR025705 Beta_hexosaminidase_sua/sub
IPR029018 Hex-like_dom2
IPR015883 Glyco_hydro_20_cat
IPR029019 HEX_eukaryotic_N
IPR036412 HAD-like_sf
IPR004274 FCP1_dom
IPR036734 Neur_chan_lig-bd_sf
IPR006202 Neur_chan_lig-bd
IPR004625 PyrdxlKinase
IPR013749 PM/HMP-P_kinase-1
IPR029056 Ribokinase-like
SUPFAM
Gene 3D
ProteinModelPortal
Q3L6N4
H9JX51
A4PHN7
Q3L6N3
H9JX50
A4LAF9
+ More
Q2Q1D3 Q3LS76 Q2Q1D2 Q3LS75 A0A194QY72 A0A212F3V9 A0A194QY01 A0A194PM72 A0A2H1V9E6 B6CI24 B0XBN5 J9HI66 Q17BL1 A0A182KFH4 A0A182QVJ5 A0A182VFS0 A0A182HR23 A0A182W8J9 A0A182L7V3 A0A1L8DQZ7 A0A0L7KE68 B0XBN6 A0A182Y637 A0A182JLR9 A0A026WT31 E0VQW8 Q7Q0Z2 U5ETX4 A0A3G5BIJ3 W5JJY2 A0A1W6EW10 A0A1B0CZC5 A0A182R1R5 D6WRR7 A0A1Y1LY98 A0A1J1HQF6 A0A087ZUU5 A0A084W3Y6 A0A0M8ZZH8 A0A2J7PCV4 A0A0L7QS85 A0A1B6CMM3 A0A2P8Y3J8 D6WRR9 A0A182PTQ9 A0A182UFT6 A0A182XHA2 D6WRR8 A0A182NNT6 A0A182GID0 U4TU30 N6T5A7 A0A2J7PCU7 A0A182FN54 A0A067R5X1 A0A1W4XBS7 V5H217 A0A182LTR5 A0A147B289 A0A1B6KR81 A0A146Z1P6 A0A3Q2QBK9 A0A1B6KBG0 A0A0C9RX26 A0A1S3HJI4 B7PE53 A0A3Q1IDG1 A0A1D2MNV3 A0A0K2SW92 A0A131YVJ8 A0A1S3HGE0 A0A1B6EVU7 A0A3Q1EKY7 A0A1S3HI83 V3ZMB9 Q7YTB2 L7MG67 A0A224YPI0 A0A3B1J6J9 A0A3P8XAC0 A0A131XKM5 A0A1S3HGG6 A0A3Q3G324 A0A3B3Q349 A0A2D0QB43 A0A3M7R7F3 A0A3B1J888 A0A3Q1CKD5 A0A3Q1H4L2 A0A2R5LAG8 A0A0A1WSN4 A0A131XIX2 A0A3Q1I5U8 A0A3B1K7J4
Q2Q1D3 Q3LS76 Q2Q1D2 Q3LS75 A0A194QY72 A0A212F3V9 A0A194QY01 A0A194PM72 A0A2H1V9E6 B6CI24 B0XBN5 J9HI66 Q17BL1 A0A182KFH4 A0A182QVJ5 A0A182VFS0 A0A182HR23 A0A182W8J9 A0A182L7V3 A0A1L8DQZ7 A0A0L7KE68 B0XBN6 A0A182Y637 A0A182JLR9 A0A026WT31 E0VQW8 Q7Q0Z2 U5ETX4 A0A3G5BIJ3 W5JJY2 A0A1W6EW10 A0A1B0CZC5 A0A182R1R5 D6WRR7 A0A1Y1LY98 A0A1J1HQF6 A0A087ZUU5 A0A084W3Y6 A0A0M8ZZH8 A0A2J7PCV4 A0A0L7QS85 A0A1B6CMM3 A0A2P8Y3J8 D6WRR9 A0A182PTQ9 A0A182UFT6 A0A182XHA2 D6WRR8 A0A182NNT6 A0A182GID0 U4TU30 N6T5A7 A0A2J7PCU7 A0A182FN54 A0A067R5X1 A0A1W4XBS7 V5H217 A0A182LTR5 A0A147B289 A0A1B6KR81 A0A146Z1P6 A0A3Q2QBK9 A0A1B6KBG0 A0A0C9RX26 A0A1S3HJI4 B7PE53 A0A3Q1IDG1 A0A1D2MNV3 A0A0K2SW92 A0A131YVJ8 A0A1S3HGE0 A0A1B6EVU7 A0A3Q1EKY7 A0A1S3HI83 V3ZMB9 Q7YTB2 L7MG67 A0A224YPI0 A0A3B1J6J9 A0A3P8XAC0 A0A131XKM5 A0A1S3HGG6 A0A3Q3G324 A0A3B3Q349 A0A2D0QB43 A0A3M7R7F3 A0A3B1J888 A0A3Q1CKD5 A0A3Q1H4L2 A0A2R5LAG8 A0A0A1WSN4 A0A131XIX2 A0A3Q1I5U8 A0A3B1K7J4
PDB
3LMY
E-value=6.07724e-90,
Score=845
Ontologies
KEGG
PATHWAY
00511
Other glycan degradation - Bombyx mori (domestic silkworm)
00520 Amino sugar and nucleotide sugar metabolism - Bombyx mori (domestic silkworm)
00531 Glycosaminoglycan degradation - Bombyx mori (domestic silkworm)
00603 Glycosphingolipid biosynthesis - globo and isoglobo series - Bombyx mori (domestic silkworm)
00604 Glycosphingolipid biosynthesis - ganglio series - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
04142 Lysosome - Bombyx mori (domestic silkworm)
00520 Amino sugar and nucleotide sugar metabolism - Bombyx mori (domestic silkworm)
00531 Glycosaminoglycan degradation - Bombyx mori (domestic silkworm)
00603 Glycosphingolipid biosynthesis - globo and isoglobo series - Bombyx mori (domestic silkworm)
00604 Glycosphingolipid biosynthesis - ganglio series - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
04142 Lysosome - Bombyx mori (domestic silkworm)
GO
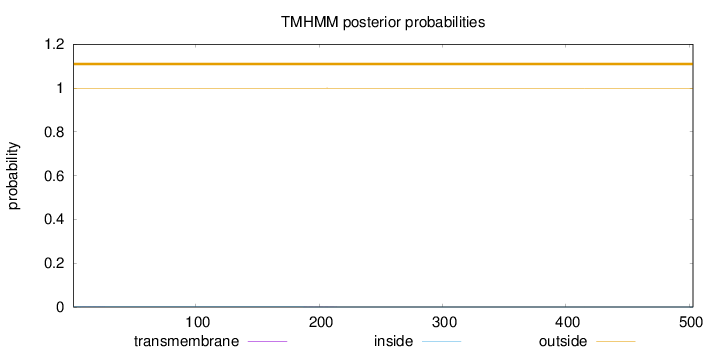
Topology
Length:
503
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01208
Exp number, first 60 AAs:
0.00516
Total prob of N-in:
0.00092
outside
1 - 503
Population Genetic Test Statistics
Pi
167.45173
Theta
153.98996
Tajima's D
0.452305
CLR
0.361197
CSRT
0.495125243737813
Interpretation
Uncertain
