Gene
KWMTBOMO04298
Pre Gene Modal
BGIBMGA014117
Annotation
PREDICTED:_dullard-like_protein_isoform_X1_[Bombyx_mori]
Full name
CTD nuclear envelope phosphatase 1 homolog
Alternative Name
Serine/threonine-protein phosphatase dullard homolog
Location in the cell
Mitochondrial Reliability : 1.731
Sequence
CDS
ATGCTGAAAAGGTTGCAAATGGGCCTTCGCGCGTTCATGCTAGTAGCATCAAAAGTATGGAGTTGTATTTGTTATATGTTTAAGAAGCAATATCGTGCGTTGGCACAATACCAATCAGTGAAGTATGAAATCTACCCACTTTCCCCAGTATCTAGACACAGGCTGAGTCTAGTTAGAAGAAAAATCCTGGTTTTAGATTTAGATGAGACATTAATCCATTCGCATCATGATGCGATGGTCAGGCCCACTGTTAAGCCGGGTACGCCTCCGGACTTTGTCCTGAAAGTCACTATCGACAAACATCCAGTTAGATTTTTTGTTCATAAGAGGCCGCATGTAGACTACTTTTTGGATATAGTTTCACAGTGGTATGAGTTGGTTGTGTTTACTGCGTCTATGGAGATTTACGGCGCCGCTGTGGCTGACAAACTAGATAATGGACGAGGTATCCTGAGGAGACGTTTTTACAGGCAACACTGTACAGCAGAACATGGATCGTCATATACTAAAAATCTGTCATCAATATGTGACGATCTGAACAGAGTGTTCATACTGGACAATTCACCTGGAGCTTACCGAGATTTCCCGGATAACGCGATTCCAATAAAATCGTGGTTCTGCGATCCGCTGGACGTGTCCCTGCTGTCGCTGCTGCCCGTGCTCGACGCGCTGCGCTTCACGCACGACGTGCGCTCCGTGCTCTCGCGCAACCTGCACCTGCACAGGCTCTGGGACCCACCGGCTTTTGTACAAATGAAGGGCGTCGCCGGCACGCCGCAGTCTATTTATTTCCCACTCGTTAGGACGTTCAACGCTTTTCAAAATATAGATAATAATTATTATACTTAA
Protein
MLKRLQMGLRAFMLVASKVWSCICYMFKKQYRALAQYQSVKYEIYPLSPVSRHRLSLVRRKILVLDLDETLIHSHHDAMVRPTVKPGTPPDFVLKVTIDKHPVRFFVHKRPHVDYFLDIVSQWYELVVFTASMEIYGAAVADKLDNGRGILRRRFYRQHCTAEHGSSYTKNLSSICDDLNRVFILDNSPGAYRDFPDNAIPIKSWFCDPLDVSLLSLLPVLDALRFTHDVRSVLSRNLHLHRLWDPPAFVQMKGVAGTPQSIYFPLVRTFNAFQNIDNNYYT
Summary
Description
Serine/threonine protein phosphatase that may dephosphorylate and activate lipin-like phosphatases. Lipins are phosphatidate phosphatases that catalyze the conversion of phosphatidic acid to diacylglycerol and control the metabolism of fatty acids at different levels. May indirectly modulate the lipid composition of nuclear and/or endoplasmic reticulum membranes and be required for proper nuclear membrane morphology and/or dynamics. May also indirectly regulate the production of lipid droplets and triacylglycerol (By similarity).
Catalytic Activity
H2O + O-phospho-L-seryl-[protein] = L-seryl-[protein] + phosphate
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
Similarity
Belongs to the dullard family.
Keywords
Complete proteome
Hydrolase
Membrane
Protein phosphatase
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain CTD nuclear envelope phosphatase 1 homolog
Uniprot
Q1HQ92
A0A1E1W038
A0A2W1BPT7
A0A2H1V110
A0A3S2N8Z4
A0A194RGW2
+ More
A0A222AJC1 H9JX52 A0A2P8YRZ2 A0A067RH29 E2BIZ4 A0A195CTH3 A0A151JNI5 A0A195B5T0 E2B029 A0A1B6GKG4 A0A1B6HTV3 A0A3L8DAC0 A0A232EM21 A0A1B6KT40 A0A1S4ELE8 K7J2Q1 A0A1Y1L048 A0A154PF11 E0VK35 A0A1Y1L054 V5IDG6 A0A1S4L4G3 B7Q367 V5HWQ3 A0A1E1XV31 A0A0C9SBB5 A0A1E1X6E1 A0A1Z5L5K0 A0A293LM45 A0A293LBS5 A0A131YNX7 A0A0C9RJ75 A0A026WKJ3 T1J4B5 A0A194Q2Y1 A0A1B6CCV0 A0A131X7B0 A0A0N0U3K6 L7MHX9 A0A1W4WM95 B4I6G4 Q9VRG7 A0A0L7R6R9 A0A0P4WG47 N6TUD6 U4TY55 B4PZ26 B3NYD0 Q29I63 A0A151JYW4 B3N0G3 A0A1W4VWT7 A0A2A3EEK2 A0A3B0K7Y6 A0A2M4CQP0 B4JLL8 B4N1H8 A0A1W4WB71 A0A1W4WLA3 E9FYK0 A0A0P5YKX9 B4M1V8 Q16T51 A0A1B6DA32 A0A0L0C260 X2JCV6 A0A1A9XX20 A0A1A9VNM3 A0A1B0A7P5 A0A0P5NHL9 A0A182UE25 A0A182LLK0 Q7Q5S6 A0A182X8K6 A0A240PP16 A0A0Q9WRS0 A0A0P8YEP3 A0A182QTL1 A0A0R3NZ59 A0A0R1ECC0 A0A0Q5TLD8 A0A1I8MLE9 A0A2M4AWW6 B4L8M8 A0A1W4VIK1 W5JEG0 A0A182FDT3 A0A212F6S5 A0A2M4AWN1 A0A1Q3G0P4 A0A1I8PF27 D6X3A1 A0A132AFB6 A0A0Q9WCE4
A0A222AJC1 H9JX52 A0A2P8YRZ2 A0A067RH29 E2BIZ4 A0A195CTH3 A0A151JNI5 A0A195B5T0 E2B029 A0A1B6GKG4 A0A1B6HTV3 A0A3L8DAC0 A0A232EM21 A0A1B6KT40 A0A1S4ELE8 K7J2Q1 A0A1Y1L048 A0A154PF11 E0VK35 A0A1Y1L054 V5IDG6 A0A1S4L4G3 B7Q367 V5HWQ3 A0A1E1XV31 A0A0C9SBB5 A0A1E1X6E1 A0A1Z5L5K0 A0A293LM45 A0A293LBS5 A0A131YNX7 A0A0C9RJ75 A0A026WKJ3 T1J4B5 A0A194Q2Y1 A0A1B6CCV0 A0A131X7B0 A0A0N0U3K6 L7MHX9 A0A1W4WM95 B4I6G4 Q9VRG7 A0A0L7R6R9 A0A0P4WG47 N6TUD6 U4TY55 B4PZ26 B3NYD0 Q29I63 A0A151JYW4 B3N0G3 A0A1W4VWT7 A0A2A3EEK2 A0A3B0K7Y6 A0A2M4CQP0 B4JLL8 B4N1H8 A0A1W4WB71 A0A1W4WLA3 E9FYK0 A0A0P5YKX9 B4M1V8 Q16T51 A0A1B6DA32 A0A0L0C260 X2JCV6 A0A1A9XX20 A0A1A9VNM3 A0A1B0A7P5 A0A0P5NHL9 A0A182UE25 A0A182LLK0 Q7Q5S6 A0A182X8K6 A0A240PP16 A0A0Q9WRS0 A0A0P8YEP3 A0A182QTL1 A0A0R3NZ59 A0A0R1ECC0 A0A0Q5TLD8 A0A1I8MLE9 A0A2M4AWW6 B4L8M8 A0A1W4VIK1 W5JEG0 A0A182FDT3 A0A212F6S5 A0A2M4AWN1 A0A1Q3G0P4 A0A1I8PF27 D6X3A1 A0A132AFB6 A0A0Q9WCE4
EC Number
3.1.3.16
Pubmed
28756777
26354079
19121390
29403074
24845553
20798317
+ More
30249741 28648823 20075255 28004739 20566863 25765539 29209593 26131772 28503490 28528879 26830274 24508170 28049606 25576852 17994087 10731132 12537572 12537569 23537049 17550304 15632085 21292972 17510324 26108605 12537568 12537573 12537574 16110336 17569856 17569867 20966253 12364791 25315136 18057021 20920257 23761445 22118469 18362917 19820115 26555130
30249741 28648823 20075255 28004739 20566863 25765539 29209593 26131772 28503490 28528879 26830274 24508170 28049606 25576852 17994087 10731132 12537572 12537569 23537049 17550304 15632085 21292972 17510324 26108605 12537568 12537573 12537574 16110336 17569856 17569867 20966253 12364791 25315136 18057021 20920257 23761445 22118469 18362917 19820115 26555130
EMBL
DQ443160
ABF51249.1
GDQN01010707
JAT80347.1
KZ150021
PZC74886.1
+ More
ODYU01000179 SOQ34535.1 RSAL01000190 RVE44704.1 KQ460211 KPJ16560.1 KY609677 ASO76454.1 BABH01031929 PYGN01000400 PSN47007.1 KK852510 KDR22343.1 GL448543 EFN84339.1 KQ977279 KYN03998.1 KQ978820 KYN28125.1 KQ976595 KYM79617.1 GL444372 EFN60955.1 GECZ01006986 JAS62783.1 GECU01029592 JAS78114.1 QOIP01000010 RLU17445.1 NNAY01003456 OXU19387.1 GEBQ01025389 JAT14588.1 GEZM01068298 GEZM01068296 GEZM01068295 JAV67032.1 KQ434890 KZC10431.1 DS235239 EEB13741.1 GEZM01068297 JAV67033.1 GANP01011972 JAB72496.1 ABJB010220444 ABJB010287002 ABJB010889996 ABJB010991774 DS847973 EEC13289.1 GANP01005570 JAB78898.1 GFAA01000361 JAU03074.1 GBZX01003062 JAG89678.1 GFAC01004404 JAT94784.1 GFJQ02004699 JAW02271.1 GFWV01005073 MAA29803.1 GFWV01000518 MAA25248.1 GEDV01008417 JAP80140.1 GBYB01013277 JAG83044.1 KK107167 EZA56503.1 JH431842 KQ459580 KPI99359.1 GEDC01026029 JAS11269.1 GEFH01005077 JAP63504.1 KQ435896 KOX69301.1 GACK01001469 JAA63565.1 CH480823 EDW56370.1 AE014298 AY094776 AY118321 KQ414646 KOC66543.1 GDRN01059520 JAI65493.1 APGK01018465 APGK01018466 APGK01018467 APGK01018468 KB740076 ENN81653.1 KB631643 ERL84923.1 CM000162 EDX03087.1 CH954180 EDV47609.1 CH379063 KQ981451 KYN41628.1 CH902640 EDV38367.2 KZ288277 PBC29712.1 OUUW01000021 SPP89443.1 GGFL01003459 MBW67637.1 CH916370 EDW00471.1 CH963925 EDW78217.1 GL732527 EFX87753.1 GDIP01057414 LRGB01001151 JAM46301.1 KZS13477.1 CH940651 EDW65662.1 CH477659 EAT37656.1 EAT37657.1 GEDC01014818 JAS22480.1 JRES01001107 KNC25509.1 AHN59985.1 GDIQ01152593 JAK99132.1 AAAB01008960 EAA10772.1 KRF98833.1 KPU77382.1 AXCN02000553 KRT06328.1 KRK07099.1 KQS30570.1 GGFK01011881 MBW45202.1 CH933815 EDW08003.1 KRG07540.1 ADMH02001746 ETN61214.1 AGBW02009971 OWR49446.1 GGFK01011882 MBW45203.1 GFDL01001678 JAV33367.1 KQ971372 EFA09780.2 JXLN01013336 KPM09255.1 KRF82332.1
ODYU01000179 SOQ34535.1 RSAL01000190 RVE44704.1 KQ460211 KPJ16560.1 KY609677 ASO76454.1 BABH01031929 PYGN01000400 PSN47007.1 KK852510 KDR22343.1 GL448543 EFN84339.1 KQ977279 KYN03998.1 KQ978820 KYN28125.1 KQ976595 KYM79617.1 GL444372 EFN60955.1 GECZ01006986 JAS62783.1 GECU01029592 JAS78114.1 QOIP01000010 RLU17445.1 NNAY01003456 OXU19387.1 GEBQ01025389 JAT14588.1 GEZM01068298 GEZM01068296 GEZM01068295 JAV67032.1 KQ434890 KZC10431.1 DS235239 EEB13741.1 GEZM01068297 JAV67033.1 GANP01011972 JAB72496.1 ABJB010220444 ABJB010287002 ABJB010889996 ABJB010991774 DS847973 EEC13289.1 GANP01005570 JAB78898.1 GFAA01000361 JAU03074.1 GBZX01003062 JAG89678.1 GFAC01004404 JAT94784.1 GFJQ02004699 JAW02271.1 GFWV01005073 MAA29803.1 GFWV01000518 MAA25248.1 GEDV01008417 JAP80140.1 GBYB01013277 JAG83044.1 KK107167 EZA56503.1 JH431842 KQ459580 KPI99359.1 GEDC01026029 JAS11269.1 GEFH01005077 JAP63504.1 KQ435896 KOX69301.1 GACK01001469 JAA63565.1 CH480823 EDW56370.1 AE014298 AY094776 AY118321 KQ414646 KOC66543.1 GDRN01059520 JAI65493.1 APGK01018465 APGK01018466 APGK01018467 APGK01018468 KB740076 ENN81653.1 KB631643 ERL84923.1 CM000162 EDX03087.1 CH954180 EDV47609.1 CH379063 KQ981451 KYN41628.1 CH902640 EDV38367.2 KZ288277 PBC29712.1 OUUW01000021 SPP89443.1 GGFL01003459 MBW67637.1 CH916370 EDW00471.1 CH963925 EDW78217.1 GL732527 EFX87753.1 GDIP01057414 LRGB01001151 JAM46301.1 KZS13477.1 CH940651 EDW65662.1 CH477659 EAT37656.1 EAT37657.1 GEDC01014818 JAS22480.1 JRES01001107 KNC25509.1 AHN59985.1 GDIQ01152593 JAK99132.1 AAAB01008960 EAA10772.1 KRF98833.1 KPU77382.1 AXCN02000553 KRT06328.1 KRK07099.1 KQS30570.1 GGFK01011881 MBW45202.1 CH933815 EDW08003.1 KRG07540.1 ADMH02001746 ETN61214.1 AGBW02009971 OWR49446.1 GGFK01011882 MBW45203.1 GFDL01001678 JAV33367.1 KQ971372 EFA09780.2 JXLN01013336 KPM09255.1 KRF82332.1
Proteomes
UP000283053
UP000053240
UP000005204
UP000245037
UP000027135
UP000008237
+ More
UP000078542 UP000078492 UP000078540 UP000000311 UP000279307 UP000215335 UP000079169 UP000002358 UP000076502 UP000009046 UP000001555 UP000053097 UP000053268 UP000053105 UP000192223 UP000001292 UP000000803 UP000053825 UP000019118 UP000030742 UP000002282 UP000008711 UP000001819 UP000078541 UP000007801 UP000192221 UP000242457 UP000268350 UP000001070 UP000007798 UP000000305 UP000076858 UP000008792 UP000008820 UP000037069 UP000092443 UP000078200 UP000092445 UP000075902 UP000075882 UP000007062 UP000076407 UP000075880 UP000075886 UP000095301 UP000009192 UP000000673 UP000069272 UP000007151 UP000095300 UP000007266
UP000078542 UP000078492 UP000078540 UP000000311 UP000279307 UP000215335 UP000079169 UP000002358 UP000076502 UP000009046 UP000001555 UP000053097 UP000053268 UP000053105 UP000192223 UP000001292 UP000000803 UP000053825 UP000019118 UP000030742 UP000002282 UP000008711 UP000001819 UP000078541 UP000007801 UP000192221 UP000242457 UP000268350 UP000001070 UP000007798 UP000000305 UP000076858 UP000008792 UP000008820 UP000037069 UP000092443 UP000078200 UP000092445 UP000075902 UP000075882 UP000007062 UP000076407 UP000075880 UP000075886 UP000095301 UP000009192 UP000000673 UP000069272 UP000007151 UP000095300 UP000007266
Pfam
PF03031 NIF
Interpro
SUPFAM
SSF56784
SSF56784
Gene 3D
ProteinModelPortal
Q1HQ92
A0A1E1W038
A0A2W1BPT7
A0A2H1V110
A0A3S2N8Z4
A0A194RGW2
+ More
A0A222AJC1 H9JX52 A0A2P8YRZ2 A0A067RH29 E2BIZ4 A0A195CTH3 A0A151JNI5 A0A195B5T0 E2B029 A0A1B6GKG4 A0A1B6HTV3 A0A3L8DAC0 A0A232EM21 A0A1B6KT40 A0A1S4ELE8 K7J2Q1 A0A1Y1L048 A0A154PF11 E0VK35 A0A1Y1L054 V5IDG6 A0A1S4L4G3 B7Q367 V5HWQ3 A0A1E1XV31 A0A0C9SBB5 A0A1E1X6E1 A0A1Z5L5K0 A0A293LM45 A0A293LBS5 A0A131YNX7 A0A0C9RJ75 A0A026WKJ3 T1J4B5 A0A194Q2Y1 A0A1B6CCV0 A0A131X7B0 A0A0N0U3K6 L7MHX9 A0A1W4WM95 B4I6G4 Q9VRG7 A0A0L7R6R9 A0A0P4WG47 N6TUD6 U4TY55 B4PZ26 B3NYD0 Q29I63 A0A151JYW4 B3N0G3 A0A1W4VWT7 A0A2A3EEK2 A0A3B0K7Y6 A0A2M4CQP0 B4JLL8 B4N1H8 A0A1W4WB71 A0A1W4WLA3 E9FYK0 A0A0P5YKX9 B4M1V8 Q16T51 A0A1B6DA32 A0A0L0C260 X2JCV6 A0A1A9XX20 A0A1A9VNM3 A0A1B0A7P5 A0A0P5NHL9 A0A182UE25 A0A182LLK0 Q7Q5S6 A0A182X8K6 A0A240PP16 A0A0Q9WRS0 A0A0P8YEP3 A0A182QTL1 A0A0R3NZ59 A0A0R1ECC0 A0A0Q5TLD8 A0A1I8MLE9 A0A2M4AWW6 B4L8M8 A0A1W4VIK1 W5JEG0 A0A182FDT3 A0A212F6S5 A0A2M4AWN1 A0A1Q3G0P4 A0A1I8PF27 D6X3A1 A0A132AFB6 A0A0Q9WCE4
A0A222AJC1 H9JX52 A0A2P8YRZ2 A0A067RH29 E2BIZ4 A0A195CTH3 A0A151JNI5 A0A195B5T0 E2B029 A0A1B6GKG4 A0A1B6HTV3 A0A3L8DAC0 A0A232EM21 A0A1B6KT40 A0A1S4ELE8 K7J2Q1 A0A1Y1L048 A0A154PF11 E0VK35 A0A1Y1L054 V5IDG6 A0A1S4L4G3 B7Q367 V5HWQ3 A0A1E1XV31 A0A0C9SBB5 A0A1E1X6E1 A0A1Z5L5K0 A0A293LM45 A0A293LBS5 A0A131YNX7 A0A0C9RJ75 A0A026WKJ3 T1J4B5 A0A194Q2Y1 A0A1B6CCV0 A0A131X7B0 A0A0N0U3K6 L7MHX9 A0A1W4WM95 B4I6G4 Q9VRG7 A0A0L7R6R9 A0A0P4WG47 N6TUD6 U4TY55 B4PZ26 B3NYD0 Q29I63 A0A151JYW4 B3N0G3 A0A1W4VWT7 A0A2A3EEK2 A0A3B0K7Y6 A0A2M4CQP0 B4JLL8 B4N1H8 A0A1W4WB71 A0A1W4WLA3 E9FYK0 A0A0P5YKX9 B4M1V8 Q16T51 A0A1B6DA32 A0A0L0C260 X2JCV6 A0A1A9XX20 A0A1A9VNM3 A0A1B0A7P5 A0A0P5NHL9 A0A182UE25 A0A182LLK0 Q7Q5S6 A0A182X8K6 A0A240PP16 A0A0Q9WRS0 A0A0P8YEP3 A0A182QTL1 A0A0R3NZ59 A0A0R1ECC0 A0A0Q5TLD8 A0A1I8MLE9 A0A2M4AWW6 B4L8M8 A0A1W4VIK1 W5JEG0 A0A182FDT3 A0A212F6S5 A0A2M4AWN1 A0A1Q3G0P4 A0A1I8PF27 D6X3A1 A0A132AFB6 A0A0Q9WCE4
PDB
2HHL
E-value=1.20529e-26,
Score=296
Ontologies
GO
Topology
Subcellular location
Membrane
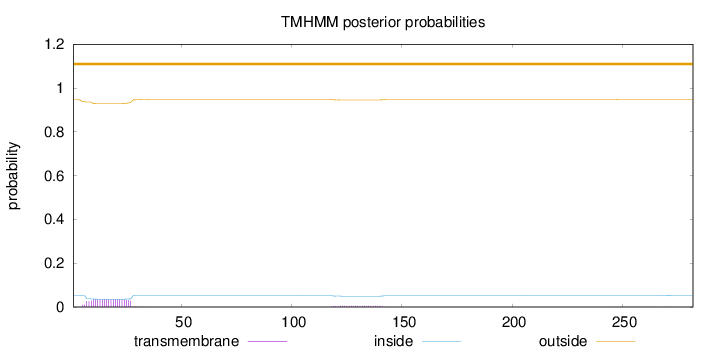
Length:
282
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.8911
Exp number, first 60 AAs:
0.738559999999999
Total prob of N-in:
0.05399
outside
1 - 282
Population Genetic Test Statistics
Pi
171.934773
Theta
175.762814
Tajima's D
-0.620409
CLR
0.666324
CSRT
0.219289035548223
Interpretation
Uncertain
