Gene
KWMTBOMO04297 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014121
Annotation
PREDICTED:_mRNA-decapping_enzyme_1A_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 2.793
Sequence
CDS
ATGGCTGACACCGGGTTACGTAGTATGAATTTCGCTGCATTAAAGCGGGCAGATCCATATGCTCGTGAAATTGTCGATAGTGCTACCCACGTTGCCCTTTACACATTTGAAGAGAACGAATGGGAGAAAACCAACATTGAGGGTGCTTTGTTTGTGTACAGTAGAAATGGTGAACCATACCACAGTTTAGTGATTATGAACAGGCTGAACACTAACAACCTAATAGAACCAGTGTCTAAAGGAATTGAACTACAGTTAAAGGAGCCGTTTTTACTATATCGTAACGCCAAGTGCCGCATTTACGGTATATGGTTTTATGATAAGGACGAGTGCGTGCGAGTGGCCACGAAGTTGAACACATTAGTGAAAGAGTCCATAAAACAGTCGCAAGGCGAAACCATGCCACAGAATTCAGTTTACAATTCGTCTACAACTTCAACTCAATCTATAGATATCTTCAGCATGTTGAGTAAAGCGCAAGACGATTTCAATTCTAATAAAGGTGTTGCTGGAAACAAATCCGAATCGACGCCGGCCAAAGCGCCTGACATGACGTCGCAAAGTGTAATGGATTTCTTTGCAAAGGCTGGGAGTGGAGCATCGGCTCAAATGCCTGCAGTGTCTTCATTGCCCTCACCAGGGATATTCGGTCCGCGTGCCACTGATTCCAGAGAAGTACCACAGCTTTTACAGAGGCTCATGAGTAACCCTGCACATTCAGTAGAACATATTGAGAAGCAACAGCGATCTGTTACTCCTCAAGAAGGGCAAAGCTCAAATGGTAATTCTACACTAGATTCTGGTATCAAACCAAATTGTATGTTCTCATTAAAGTCAACTCCTTTGGAGAAGCAAAATCAAAGGATACCAGGATTGAAAAAAGCTCTGGATAGTAGTGATGCTAATGGAATAGTCACCATGGAGAATGAACTCAATTTGATGCACTTATCTTCTCCCAAGACAACATCGCCATTAGCTGCATACTTGAATCAATCTCAAGACATAGGACACACGAAACCGGCGCTGATGCCCCCCACAATGTTCACTGCAAGTTCAGGTGGTGATGTCCAGCCATCACCGCAGCCACTCACCAGGAATCAACTCCTTCAGGCCTTTAATTACTTACTCAAACATGATGCAGATTTTGTGAATAAACTTCACGAAGCCTATGTCAAATCGTTTTCAGAAAAAGCATTTTCTGTGTCATAA
Protein
MADTGLRSMNFAALKRADPYAREIVDSATHVALYTFEENEWEKTNIEGALFVYSRNGEPYHSLVIMNRLNTNNLIEPVSKGIELQLKEPFLLYRNAKCRIYGIWFYDKDECVRVATKLNTLVKESIKQSQGETMPQNSVYNSSTTSTQSIDIFSMLSKAQDDFNSNKGVAGNKSESTPAKAPDMTSQSVMDFFAKAGSGASAQMPAVSSLPSPGIFGPRATDSREVPQLLQRLMSNPAHSVEHIEKQQRSVTPQEGQSSNGNSTLDSGIKPNCMFSLKSTPLEKQNQRIPGLKKALDSSDANGIVTMENELNLMHLSSPKTTSPLAAYLNQSQDIGHTKPALMPPTMFTASSGGDVQPSPQPLTRNQLLQAFNYLLKHDADFVNKLHEAYVKSFSEKAFSVS
Summary
Uniprot
H9JX56
A0A2A4IV98
A0A2W1BNM9
A0A2H1V1A5
A0A194Q1Z9
A0A194RKR5
+ More
S4NSU2 A0A212F6T8 A0A0L7LDK1 A0A067R8W4 A0A195B614 A0A158NQA7 A0A151JNJ4 A0A2J7PTV7 A0A151X9Y4 A0A154PF41 A0A151K116 A0A0T6AVZ9 A0A0L7R6M3 F4WBE9 E2B027 A0A3L8DC29 A0A026WLH7 A0A195CV03 E2BEK1 A0A0N0U393 A0A088A9F6 A0A2A3ED57 A0A1B6I4A3 A0A1B6GVX2 A0A2P8XI78 A0A1B6MRT8 A0A1B0GLW1 A0A1W4XLQ9 A0A1B6CJ42 A0A1L8DFR7 A0A1Y1NJX3 A0A1L8DGY6 A0A1B0CJA2 D6X0J0 E0VZ25 U4UME1 N6TM53 J9K8Y4 A0A2S2Q3R3 A0A1S3CW67 B0WEB0 Q17M13 A0A1S4EY16 A0A1Q3F0L5 A0A0K8TLC4 A0NAU2 A0A1Q3F002 A0A3B0JIU8 A0A034V8R5 A0A0K8UPI3 A0A336LMB7 A0A0A1WHN9 A0A1I8PGD1 B4KMC8 A0A0L0C9F6 A0A2R7VR28 A0A0M3QVL6 B4LNY7 B4GCX9 B4J643 Q292R0 B4MRP3 B3NQ15 W8CBP9 B4I8V6 B4QB16 B4PA91 A0A182V4G3 B3MCL7 T1H0H7 Q9W1H5 Q52KA4 A0A182GF77 A0A1I8N5D9 A0A182JFE2 A0A023EU20 A0A182GHH4 A0A1W4UMK9 A0A1J1IHV5 A0A1A9WP54 A0A1A9Y6H9 A0A1B0BI89 A0A0P6DTF4 A0A0N8C3I2 A0A0N8EC61 A0A0N8CBQ2 A0A1A9UJ05 A0A182NF54
S4NSU2 A0A212F6T8 A0A0L7LDK1 A0A067R8W4 A0A195B614 A0A158NQA7 A0A151JNJ4 A0A2J7PTV7 A0A151X9Y4 A0A154PF41 A0A151K116 A0A0T6AVZ9 A0A0L7R6M3 F4WBE9 E2B027 A0A3L8DC29 A0A026WLH7 A0A195CV03 E2BEK1 A0A0N0U393 A0A088A9F6 A0A2A3ED57 A0A1B6I4A3 A0A1B6GVX2 A0A2P8XI78 A0A1B6MRT8 A0A1B0GLW1 A0A1W4XLQ9 A0A1B6CJ42 A0A1L8DFR7 A0A1Y1NJX3 A0A1L8DGY6 A0A1B0CJA2 D6X0J0 E0VZ25 U4UME1 N6TM53 J9K8Y4 A0A2S2Q3R3 A0A1S3CW67 B0WEB0 Q17M13 A0A1S4EY16 A0A1Q3F0L5 A0A0K8TLC4 A0NAU2 A0A1Q3F002 A0A3B0JIU8 A0A034V8R5 A0A0K8UPI3 A0A336LMB7 A0A0A1WHN9 A0A1I8PGD1 B4KMC8 A0A0L0C9F6 A0A2R7VR28 A0A0M3QVL6 B4LNY7 B4GCX9 B4J643 Q292R0 B4MRP3 B3NQ15 W8CBP9 B4I8V6 B4QB16 B4PA91 A0A182V4G3 B3MCL7 T1H0H7 Q9W1H5 Q52KA4 A0A182GF77 A0A1I8N5D9 A0A182JFE2 A0A023EU20 A0A182GHH4 A0A1W4UMK9 A0A1J1IHV5 A0A1A9WP54 A0A1A9Y6H9 A0A1B0BI89 A0A0P6DTF4 A0A0N8C3I2 A0A0N8EC61 A0A0N8CBQ2 A0A1A9UJ05 A0A182NF54
Pubmed
19121390
28756777
26354079
23622113
22118469
26227816
+ More
24845553 21347285 21719571 20798317 30249741 24508170 29403074 28004739 18362917 19820115 20566863 23537049 17510324 26369729 9087549 12364791 14747013 17210077 25348373 25830018 17994087 26108605 15632085 24495485 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19966221 23142987 26109357 26109356 26483478 25315136 24945155
24845553 21347285 21719571 20798317 30249741 24508170 29403074 28004739 18362917 19820115 20566863 23537049 17510324 26369729 9087549 12364791 14747013 17210077 25348373 25830018 17994087 26108605 15632085 24495485 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19966221 23142987 26109357 26109356 26483478 25315136 24945155
EMBL
BABH01031927
NWSH01006116
PCG63651.1
KZ150021
PZC74887.1
ODYU01000179
+ More
SOQ34536.1 KQ459580 KPI99358.1 KQ460211 KPJ16561.1 GAIX01013912 JAA78648.1 AGBW02009971 OWR49447.1 JTDY01001552 KOB73557.1 KK852619 KDR20096.1 KQ976595 KYM79619.1 ADTU01023008 KQ978820 KYN28127.1 NEVH01021221 PNF19761.1 KQ982365 KYQ57120.1 KQ434890 KZC10432.1 KQ981189 KYN45215.1 LJIG01022737 KRT78974.1 KQ414646 KOC66542.1 GL888062 EGI68505.1 GL444372 EFN60953.1 QOIP01000010 RLU17439.1 KK107167 EZA56506.1 KQ977279 KYN03994.1 GL447817 EFN85891.1 KQ435896 KOX69302.1 KZ288277 PBC29713.1 GECU01025935 JAS81771.1 GECZ01003276 JAS66493.1 PYGN01002041 PSN31715.1 GEBQ01020398 GEBQ01001368 JAT19579.1 JAT38609.1 AJVK01009138 AJVK01009139 AJVK01009140 GEDC01023868 GEDC01021424 GEDC01005759 GEDC01000834 JAS13430.1 JAS15874.1 JAS31539.1 JAS36464.1 GFDF01008798 JAV05286.1 GEZM01001061 JAV98191.1 GFDF01008480 JAV05604.1 AJWK01014365 KQ971371 EFA10020.1 DS235849 EEB18631.1 KB632276 ERL91175.1 APGK01058895 APGK01058896 KB741292 ENN70335.1 ABLF02030669 GGMS01003153 MBY72356.1 DS231907 EDS45470.1 CH477209 EAT47731.1 GFDL01013961 JAV21084.1 GDAI01002479 JAI15124.1 AAAB01006459 EAU77895.2 GFDL01014171 JAV20874.1 OUUW01000001 SPP73166.1 GAKP01020812 JAC38140.1 GDHF01023700 JAI28614.1 UFQT01000021 SSX17911.1 GBXI01015723 JAC98568.1 CH933808 EDW09816.1 JRES01001518 JRES01000737 KNC22293.1 KNC28867.1 KK854031 PTY09856.1 CP012524 ALC42619.1 CH940648 EDW61156.1 CH479181 EDW31517.1 CH916367 EDW01901.1 CM000071 EAL24801.2 CH963850 EDW74782.1 CH954179 EDV56888.1 GAMC01000299 JAC06257.1 CH480824 EDW57031.1 CM000362 CM002911 EDX08453.1 KMY96149.1 CM000158 EDW92417.1 CH902619 EDV36251.1 CAQQ02373170 AE013599 AAF47089.1 ACZ94554.1 BT021964 AAX94785.1 JXUM01059531 KQ562059 KXJ76786.1 AXCP01007770 GAPW01001192 JAC12406.1 JXUM01063734 KQ562265 KXJ76272.1 CVRI01000053 CRK99784.1 JXJN01014862 GDIQ01073162 JAN21575.1 GDIQ01137242 GDIQ01118403 JAL33323.1 GDIQ01041180 JAN53557.1 GDIQ01095443 JAL56283.1
SOQ34536.1 KQ459580 KPI99358.1 KQ460211 KPJ16561.1 GAIX01013912 JAA78648.1 AGBW02009971 OWR49447.1 JTDY01001552 KOB73557.1 KK852619 KDR20096.1 KQ976595 KYM79619.1 ADTU01023008 KQ978820 KYN28127.1 NEVH01021221 PNF19761.1 KQ982365 KYQ57120.1 KQ434890 KZC10432.1 KQ981189 KYN45215.1 LJIG01022737 KRT78974.1 KQ414646 KOC66542.1 GL888062 EGI68505.1 GL444372 EFN60953.1 QOIP01000010 RLU17439.1 KK107167 EZA56506.1 KQ977279 KYN03994.1 GL447817 EFN85891.1 KQ435896 KOX69302.1 KZ288277 PBC29713.1 GECU01025935 JAS81771.1 GECZ01003276 JAS66493.1 PYGN01002041 PSN31715.1 GEBQ01020398 GEBQ01001368 JAT19579.1 JAT38609.1 AJVK01009138 AJVK01009139 AJVK01009140 GEDC01023868 GEDC01021424 GEDC01005759 GEDC01000834 JAS13430.1 JAS15874.1 JAS31539.1 JAS36464.1 GFDF01008798 JAV05286.1 GEZM01001061 JAV98191.1 GFDF01008480 JAV05604.1 AJWK01014365 KQ971371 EFA10020.1 DS235849 EEB18631.1 KB632276 ERL91175.1 APGK01058895 APGK01058896 KB741292 ENN70335.1 ABLF02030669 GGMS01003153 MBY72356.1 DS231907 EDS45470.1 CH477209 EAT47731.1 GFDL01013961 JAV21084.1 GDAI01002479 JAI15124.1 AAAB01006459 EAU77895.2 GFDL01014171 JAV20874.1 OUUW01000001 SPP73166.1 GAKP01020812 JAC38140.1 GDHF01023700 JAI28614.1 UFQT01000021 SSX17911.1 GBXI01015723 JAC98568.1 CH933808 EDW09816.1 JRES01001518 JRES01000737 KNC22293.1 KNC28867.1 KK854031 PTY09856.1 CP012524 ALC42619.1 CH940648 EDW61156.1 CH479181 EDW31517.1 CH916367 EDW01901.1 CM000071 EAL24801.2 CH963850 EDW74782.1 CH954179 EDV56888.1 GAMC01000299 JAC06257.1 CH480824 EDW57031.1 CM000362 CM002911 EDX08453.1 KMY96149.1 CM000158 EDW92417.1 CH902619 EDV36251.1 CAQQ02373170 AE013599 AAF47089.1 ACZ94554.1 BT021964 AAX94785.1 JXUM01059531 KQ562059 KXJ76786.1 AXCP01007770 GAPW01001192 JAC12406.1 JXUM01063734 KQ562265 KXJ76272.1 CVRI01000053 CRK99784.1 JXJN01014862 GDIQ01073162 JAN21575.1 GDIQ01137242 GDIQ01118403 JAL33323.1 GDIQ01041180 JAN53557.1 GDIQ01095443 JAL56283.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000027135 UP000078540 UP000005205 UP000078492 UP000235965 UP000075809 UP000076502 UP000078541 UP000053825 UP000007755 UP000000311 UP000279307 UP000053097 UP000078542 UP000008237 UP000053105 UP000005203 UP000242457 UP000245037 UP000092462 UP000192223 UP000092461 UP000007266 UP000009046 UP000030742 UP000019118 UP000007819 UP000079169 UP000002320 UP000008820 UP000007062 UP000268350 UP000095300 UP000009192 UP000037069 UP000092553 UP000008792 UP000008744 UP000001070 UP000001819 UP000007798 UP000008711 UP000001292 UP000000304 UP000002282 UP000075903 UP000007801 UP000015102 UP000000803 UP000069940 UP000249989 UP000095301 UP000075880 UP000192221 UP000183832 UP000091820 UP000092443 UP000092460 UP000078200 UP000075884
UP000027135 UP000078540 UP000005205 UP000078492 UP000235965 UP000075809 UP000076502 UP000078541 UP000053825 UP000007755 UP000000311 UP000279307 UP000053097 UP000078542 UP000008237 UP000053105 UP000005203 UP000242457 UP000245037 UP000092462 UP000192223 UP000092461 UP000007266 UP000009046 UP000030742 UP000019118 UP000007819 UP000079169 UP000002320 UP000008820 UP000007062 UP000268350 UP000095300 UP000009192 UP000037069 UP000092553 UP000008792 UP000008744 UP000001070 UP000001819 UP000007798 UP000008711 UP000001292 UP000000304 UP000002282 UP000075903 UP000007801 UP000015102 UP000000803 UP000069940 UP000249989 UP000095301 UP000075880 UP000192221 UP000183832 UP000091820 UP000092443 UP000092460 UP000078200 UP000075884
Gene 3D
ProteinModelPortal
H9JX56
A0A2A4IV98
A0A2W1BNM9
A0A2H1V1A5
A0A194Q1Z9
A0A194RKR5
+ More
S4NSU2 A0A212F6T8 A0A0L7LDK1 A0A067R8W4 A0A195B614 A0A158NQA7 A0A151JNJ4 A0A2J7PTV7 A0A151X9Y4 A0A154PF41 A0A151K116 A0A0T6AVZ9 A0A0L7R6M3 F4WBE9 E2B027 A0A3L8DC29 A0A026WLH7 A0A195CV03 E2BEK1 A0A0N0U393 A0A088A9F6 A0A2A3ED57 A0A1B6I4A3 A0A1B6GVX2 A0A2P8XI78 A0A1B6MRT8 A0A1B0GLW1 A0A1W4XLQ9 A0A1B6CJ42 A0A1L8DFR7 A0A1Y1NJX3 A0A1L8DGY6 A0A1B0CJA2 D6X0J0 E0VZ25 U4UME1 N6TM53 J9K8Y4 A0A2S2Q3R3 A0A1S3CW67 B0WEB0 Q17M13 A0A1S4EY16 A0A1Q3F0L5 A0A0K8TLC4 A0NAU2 A0A1Q3F002 A0A3B0JIU8 A0A034V8R5 A0A0K8UPI3 A0A336LMB7 A0A0A1WHN9 A0A1I8PGD1 B4KMC8 A0A0L0C9F6 A0A2R7VR28 A0A0M3QVL6 B4LNY7 B4GCX9 B4J643 Q292R0 B4MRP3 B3NQ15 W8CBP9 B4I8V6 B4QB16 B4PA91 A0A182V4G3 B3MCL7 T1H0H7 Q9W1H5 Q52KA4 A0A182GF77 A0A1I8N5D9 A0A182JFE2 A0A023EU20 A0A182GHH4 A0A1W4UMK9 A0A1J1IHV5 A0A1A9WP54 A0A1A9Y6H9 A0A1B0BI89 A0A0P6DTF4 A0A0N8C3I2 A0A0N8EC61 A0A0N8CBQ2 A0A1A9UJ05 A0A182NF54
S4NSU2 A0A212F6T8 A0A0L7LDK1 A0A067R8W4 A0A195B614 A0A158NQA7 A0A151JNJ4 A0A2J7PTV7 A0A151X9Y4 A0A154PF41 A0A151K116 A0A0T6AVZ9 A0A0L7R6M3 F4WBE9 E2B027 A0A3L8DC29 A0A026WLH7 A0A195CV03 E2BEK1 A0A0N0U393 A0A088A9F6 A0A2A3ED57 A0A1B6I4A3 A0A1B6GVX2 A0A2P8XI78 A0A1B6MRT8 A0A1B0GLW1 A0A1W4XLQ9 A0A1B6CJ42 A0A1L8DFR7 A0A1Y1NJX3 A0A1L8DGY6 A0A1B0CJA2 D6X0J0 E0VZ25 U4UME1 N6TM53 J9K8Y4 A0A2S2Q3R3 A0A1S3CW67 B0WEB0 Q17M13 A0A1S4EY16 A0A1Q3F0L5 A0A0K8TLC4 A0NAU2 A0A1Q3F002 A0A3B0JIU8 A0A034V8R5 A0A0K8UPI3 A0A336LMB7 A0A0A1WHN9 A0A1I8PGD1 B4KMC8 A0A0L0C9F6 A0A2R7VR28 A0A0M3QVL6 B4LNY7 B4GCX9 B4J643 Q292R0 B4MRP3 B3NQ15 W8CBP9 B4I8V6 B4QB16 B4PA91 A0A182V4G3 B3MCL7 T1H0H7 Q9W1H5 Q52KA4 A0A182GF77 A0A1I8N5D9 A0A182JFE2 A0A023EU20 A0A182GHH4 A0A1W4UMK9 A0A1J1IHV5 A0A1A9WP54 A0A1A9Y6H9 A0A1B0BI89 A0A0P6DTF4 A0A0N8C3I2 A0A0N8EC61 A0A0N8CBQ2 A0A1A9UJ05 A0A182NF54
PDB
2LYD
E-value=5.94001e-38,
Score=395
Ontologies
GO
PANTHER
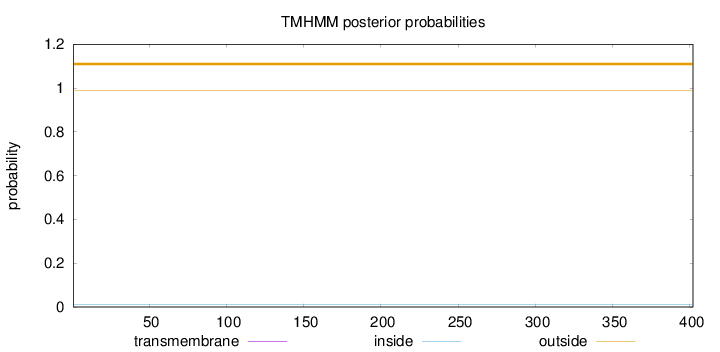
Topology
Length:
402
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00038
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01039
outside
1 - 402
Population Genetic Test Statistics
Pi
204.084693
Theta
142.079348
Tajima's D
0.079348
CLR
0
CSRT
0.390230488475576
Interpretation
Uncertain
