Gene
KWMTBOMO04291 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014573
Annotation
PREDICTED:_ankyrin_repeat_domain-containing_protein_13D-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.46
Sequence
CDS
ATGATACACAGGAACAAAAGCGGCATCTGGGGCTGGCGGCAAGACAAGACCGAGCCAGTCAACGGGTACGAGTGCAAGGTGTTCAGCGCCAATAACGTGGAGCTCGTCTCCAAGACTCGCACCGAGCATCTGGACAAGGCGCGGCGCTCCAGCCCCCGGGCTCCGCTGGCGGGGCTGCTGGCGCTCGCGGACTCCGACACCAGCGCGCCCTCTACCCCGCTCCTCACTCCTGAAACGGAAGAGCCTCCGCGCAGTAGGTCCCGTGAAGAGTTGAATGTGGTATCCTGGGAGGAGTACTTCTCTGGGGAGGCGATGGAGAGGGACATCGGGAGGCCGAAGGAGATGACCGAGAAAGTGCAAAAGTTCAAAGCCACTCTTTGGTTGTGCGAAGACTATCCATTGGAGCTGCAAGAGCAAATAATGCCGATATTGGATTTGATGGCCGCTATATCCTCGCCTCACTTCGCGAAGCTGAAGGACTTCATACAGATGCAGCTCCCGGCTGGGTTCCCAGTTAAAATAGAGATCCCGCTGTTCCACGTGCTGAACGCGCGCATCACGTTCGGCAACATCTTCGGCACGGAGAGCGGCGCGGCGCACGTGCAGTGCATCGCGGAGGGGCGGCGCGCGGCGTGCGTGCTGGACGACGCGTGCTTCGAGCTGGGCCGCGGCTACCGGGACGCCGCCGCGCCCGAGCCGCGCGGGGAGATGGACCACGACGAGCGCCTCATGCACTACGCCATACAGCAGAGCCTCATGGACGCCGGCACGCACGACGACCAGGTGGACGTGTGGGAGGCGCTGCGGGGCGCGCGGCCCCCCTCGCCGCTGCACGAGACGCAACTACAGAGGTACGAACGCTGTTATATCTTATCTTATCTTATATCTTTAAACGAGCAATTCTTGTATATATAA
Protein
MIHRNKSGIWGWRQDKTEPVNGYECKVFSANNVELVSKTRTEHLDKARRSSPRAPLAGLLALADSDTSAPSTPLLTPETEEPPRSRSREELNVVSWEEYFSGEAMERDIGRPKEMTEKVQKFKATLWLCEDYPLELQEQIMPILDLMAAISSPHFAKLKDFIQMQLPAGFPVKIEIPLFHVLNARITFGNIFGTESGAAHVQCIAEGRRAACVLDDACFELGRGYRDAAAPEPRGEMDHDERLMHYAIQQSLMDAGTHDDQVDVWEALRGARPPSPLHETQLQRYERCYILSYLISLNEQFLYI
Summary
Uniprot
H9JYF3
A0A194RKS0
A0A1E1WBA0
A0A3S2LNI4
A0A2W1BKT4
A0A212F6U0
+ More
A0A194Q7H5 A0A2A4IUT9 A0A026W2C9 A0A067R6S1 F4WKQ7 A0A0C9QNM6 A0A2A3E3B3 A0A151JZB1 A0A158NJ94 A0A195BKS7 A0A0C9PMC2 A0A0C9R620 D6WXZ2 E2AXC9 E2BSX3 A0A2J7PJU1 A0A195C3J9 A0A3L8DIY2 A0A1B6H8E1 K7IVV7 A0A0L7R8M7 A0A2J7PJT9 A0A232EE76 A0A1B6FWR0 A0A088A6I0 A0A154PET4 V5GN45 A0A2P6KX80 A0A069DZB8 A0A1B6CRZ0 A0A0P5LKM1 A0A1B6CP52 A0A0M8ZPK8 A0A1B6ECR4 A0A0P5D486 A0A224XLN8 A0A0P5WDV9 A0A0N8CP13 A0A0P6D0J0 A0A0V0GAG1 N6T8M3 A0A023F330 A0A0P5KRH1 A0A0N8D6G2 A0A0P5BGE6 A0A0P5ZEY7 A0A0P5GG21 A0A0P5KYR8 A0A0N8AUK3 A0A0P4ZW21 A0A0P5SHC8 A0A0P5K063 A0A0P5Y1W8 A0A0N8AU28 A0A0N8AV46 A0A0T6AYA9 A0A0P5BF77 A0A0P5U5N9 A0A0N8ATM7 A0A0P4XLU3 A0A0P5WUR3 A0A0P5E596 A0A3R7MMR5 A0A0P6I0I6 A0A0P5D4H6 A0A0P4XKG6 A0A0P4ZVA1 A0A0P5D4R5 A0A0P5VYG8 A0A0N7ZR06 A0A0P5Y1P5 A0A0P5SJD9 A0A0P6DJV7 A0A0P5FLX7 A0A0P6AZP8 A0A0P4Y5N2 A0A0P4Z2X2 A0A0N8E4S1 A0A0P5J336 A0A0N8ADN7 A0A0P5S613 A0A0P5KLE6 A0A1B0GJ11 A0A0P6ACY1 A0A0P5L3A3 A0A1L8DTU1 A0A0N7ZRF1 A0A0P5V8D9 A0A0P4YZ90 A0A0P5JCA0 A0A0P5ZYL2 A0A0P5SKI4
A0A194Q7H5 A0A2A4IUT9 A0A026W2C9 A0A067R6S1 F4WKQ7 A0A0C9QNM6 A0A2A3E3B3 A0A151JZB1 A0A158NJ94 A0A195BKS7 A0A0C9PMC2 A0A0C9R620 D6WXZ2 E2AXC9 E2BSX3 A0A2J7PJU1 A0A195C3J9 A0A3L8DIY2 A0A1B6H8E1 K7IVV7 A0A0L7R8M7 A0A2J7PJT9 A0A232EE76 A0A1B6FWR0 A0A088A6I0 A0A154PET4 V5GN45 A0A2P6KX80 A0A069DZB8 A0A1B6CRZ0 A0A0P5LKM1 A0A1B6CP52 A0A0M8ZPK8 A0A1B6ECR4 A0A0P5D486 A0A224XLN8 A0A0P5WDV9 A0A0N8CP13 A0A0P6D0J0 A0A0V0GAG1 N6T8M3 A0A023F330 A0A0P5KRH1 A0A0N8D6G2 A0A0P5BGE6 A0A0P5ZEY7 A0A0P5GG21 A0A0P5KYR8 A0A0N8AUK3 A0A0P4ZW21 A0A0P5SHC8 A0A0P5K063 A0A0P5Y1W8 A0A0N8AU28 A0A0N8AV46 A0A0T6AYA9 A0A0P5BF77 A0A0P5U5N9 A0A0N8ATM7 A0A0P4XLU3 A0A0P5WUR3 A0A0P5E596 A0A3R7MMR5 A0A0P6I0I6 A0A0P5D4H6 A0A0P4XKG6 A0A0P4ZVA1 A0A0P5D4R5 A0A0P5VYG8 A0A0N7ZR06 A0A0P5Y1P5 A0A0P5SJD9 A0A0P6DJV7 A0A0P5FLX7 A0A0P6AZP8 A0A0P4Y5N2 A0A0P4Z2X2 A0A0N8E4S1 A0A0P5J336 A0A0N8ADN7 A0A0P5S613 A0A0P5KLE6 A0A1B0GJ11 A0A0P6ACY1 A0A0P5L3A3 A0A1L8DTU1 A0A0N7ZRF1 A0A0P5V8D9 A0A0P4YZ90 A0A0P5JCA0 A0A0P5ZYL2 A0A0P5SKI4
Pubmed
EMBL
BABH01043411
KQ460211
KPJ16566.1
GDQN01006913
JAT84141.1
RSAL01000048
+ More
RVE50466.1 KZ150021 PZC74891.1 AGBW02009971 OWR49450.1 KQ459580 KPI99355.1 NWSH01006751 PCG63266.1 KK107536 EZA49189.1 KK852704 KDR18063.1 GL888206 EGI65149.1 GBYB01002227 JAG71994.1 KZ288405 PBC26188.1 KQ981427 KYN41770.1 ADTU01017593 KQ976453 KYM85374.1 GBYB01002228 JAG71995.1 GBYB01002226 JAG71993.1 KQ971362 EFA08921.1 GL443548 EFN61909.1 GL450311 EFN81198.1 NEVH01024944 PNF16606.1 KQ978317 KYM95175.1 QOIP01000008 RLU19808.1 GECU01036767 JAS70939.1 KQ414632 KOC67225.1 PNF16605.1 NNAY01005456 OXU16666.1 GECZ01015133 JAS54636.1 KQ434874 KZC09740.1 GALX01002937 JAB65529.1 MWRG01003984 PRD30938.1 GBGD01000915 JAC87974.1 GEDC01021110 GEDC01009121 JAS16188.1 JAS28177.1 GDIQ01172257 JAK79468.1 GEDC01022029 JAS15269.1 KQ435922 KOX68520.1 GEDC01020952 GEDC01016420 GEDC01011422 GEDC01008754 GEDC01002673 GEDC01001559 GEDC01001198 JAS16346.1 JAS20878.1 JAS25876.1 JAS28544.1 JAS34625.1 JAS35739.1 JAS36100.1 GDIP01165636 JAJ57766.1 GFTR01007363 JAW09063.1 GDIP01087446 JAM16269.1 GDIP01112943 JAL90771.1 GDIQ01084754 JAN09983.1 GECL01001203 JAP04921.1 APGK01040123 KB740975 KB632267 ENN76564.1 ERL91009.1 GBBI01003069 JAC15643.1 GDIQ01187550 JAK64175.1 GDIP01064151 JAM39564.1 GDIP01184728 JAJ38674.1 GDIP01044594 JAM59121.1 GDIQ01241436 JAK10289.1 GDIQ01182360 JAK69365.1 GDIQ01241435 JAK10290.1 GDIP01211634 JAJ11768.1 GDIP01139738 JAL63976.1 GDIQ01192808 JAK58917.1 GDIP01064150 JAM39565.1 GDIQ01242835 JAK08890.1 GDIQ01239891 JAK11834.1 LJIG01022525 KRT80121.1 GDIP01200581 JAJ22821.1 GDIP01119485 JAL84229.1 GDIQ01244043 JAK07682.1 GDIP01240027 JAI83374.1 GDIP01093385 JAM10330.1 GDIP01164887 JAJ58515.1 QCYY01003054 ROT65692.1 GDIQ01036363 JAN58374.1 GDIP01163032 JAJ60370.1 GDIP01240026 JAI83375.1 GDIP01221088 JAJ02314.1 GDIP01166428 JAJ56974.1 GDIP01093384 JAM10331.1 GDIP01221089 JAJ02313.1 GDIP01064149 JAM39566.1 GDIQ01092448 JAL59278.1 GDIQ01075791 JAN18946.1 GDIQ01253498 GDIQ01244041 GDIQ01205509 GDIQ01197953 GDIQ01196451 GDIQ01033819 JAJ98226.1 GDIP01036105 JAM67610.1 GDIP01232431 JAI90970.1 GDIP01219928 JAJ03474.1 GDIQ01061900 JAN32837.1 GDIQ01230133 JAK21592.1 GDIP01157641 JAJ65761.1 GDIQ01092449 JAL59277.1 GDIQ01182359 JAK69366.1 AJWK01019191 AJWK01019192 GDIP01036104 JAM67611.1 GDIQ01200556 JAK51169.1 GFDF01004226 JAV09858.1 GDIP01219929 JAJ03473.1 GDIP01102934 JAM00781.1 GDIP01219930 JAJ03472.1 GDIQ01244042 GDIQ01203857 JAK47868.1 GDIP01037259 JAM66456.1 GDIP01138638 JAL65076.1
RVE50466.1 KZ150021 PZC74891.1 AGBW02009971 OWR49450.1 KQ459580 KPI99355.1 NWSH01006751 PCG63266.1 KK107536 EZA49189.1 KK852704 KDR18063.1 GL888206 EGI65149.1 GBYB01002227 JAG71994.1 KZ288405 PBC26188.1 KQ981427 KYN41770.1 ADTU01017593 KQ976453 KYM85374.1 GBYB01002228 JAG71995.1 GBYB01002226 JAG71993.1 KQ971362 EFA08921.1 GL443548 EFN61909.1 GL450311 EFN81198.1 NEVH01024944 PNF16606.1 KQ978317 KYM95175.1 QOIP01000008 RLU19808.1 GECU01036767 JAS70939.1 KQ414632 KOC67225.1 PNF16605.1 NNAY01005456 OXU16666.1 GECZ01015133 JAS54636.1 KQ434874 KZC09740.1 GALX01002937 JAB65529.1 MWRG01003984 PRD30938.1 GBGD01000915 JAC87974.1 GEDC01021110 GEDC01009121 JAS16188.1 JAS28177.1 GDIQ01172257 JAK79468.1 GEDC01022029 JAS15269.1 KQ435922 KOX68520.1 GEDC01020952 GEDC01016420 GEDC01011422 GEDC01008754 GEDC01002673 GEDC01001559 GEDC01001198 JAS16346.1 JAS20878.1 JAS25876.1 JAS28544.1 JAS34625.1 JAS35739.1 JAS36100.1 GDIP01165636 JAJ57766.1 GFTR01007363 JAW09063.1 GDIP01087446 JAM16269.1 GDIP01112943 JAL90771.1 GDIQ01084754 JAN09983.1 GECL01001203 JAP04921.1 APGK01040123 KB740975 KB632267 ENN76564.1 ERL91009.1 GBBI01003069 JAC15643.1 GDIQ01187550 JAK64175.1 GDIP01064151 JAM39564.1 GDIP01184728 JAJ38674.1 GDIP01044594 JAM59121.1 GDIQ01241436 JAK10289.1 GDIQ01182360 JAK69365.1 GDIQ01241435 JAK10290.1 GDIP01211634 JAJ11768.1 GDIP01139738 JAL63976.1 GDIQ01192808 JAK58917.1 GDIP01064150 JAM39565.1 GDIQ01242835 JAK08890.1 GDIQ01239891 JAK11834.1 LJIG01022525 KRT80121.1 GDIP01200581 JAJ22821.1 GDIP01119485 JAL84229.1 GDIQ01244043 JAK07682.1 GDIP01240027 JAI83374.1 GDIP01093385 JAM10330.1 GDIP01164887 JAJ58515.1 QCYY01003054 ROT65692.1 GDIQ01036363 JAN58374.1 GDIP01163032 JAJ60370.1 GDIP01240026 JAI83375.1 GDIP01221088 JAJ02314.1 GDIP01166428 JAJ56974.1 GDIP01093384 JAM10331.1 GDIP01221089 JAJ02313.1 GDIP01064149 JAM39566.1 GDIQ01092448 JAL59278.1 GDIQ01075791 JAN18946.1 GDIQ01253498 GDIQ01244041 GDIQ01205509 GDIQ01197953 GDIQ01196451 GDIQ01033819 JAJ98226.1 GDIP01036105 JAM67610.1 GDIP01232431 JAI90970.1 GDIP01219928 JAJ03474.1 GDIQ01061900 JAN32837.1 GDIQ01230133 JAK21592.1 GDIP01157641 JAJ65761.1 GDIQ01092449 JAL59277.1 GDIQ01182359 JAK69366.1 AJWK01019191 AJWK01019192 GDIP01036104 JAM67611.1 GDIQ01200556 JAK51169.1 GFDF01004226 JAV09858.1 GDIP01219929 JAJ03473.1 GDIP01102934 JAM00781.1 GDIP01219930 JAJ03472.1 GDIQ01244042 GDIQ01203857 JAK47868.1 GDIP01037259 JAM66456.1 GDIP01138638 JAL65076.1
Proteomes
UP000005204
UP000053240
UP000283053
UP000007151
UP000053268
UP000218220
+ More
UP000053097 UP000027135 UP000007755 UP000242457 UP000078541 UP000005205 UP000078540 UP000007266 UP000000311 UP000008237 UP000235965 UP000078542 UP000279307 UP000002358 UP000053825 UP000215335 UP000005203 UP000076502 UP000053105 UP000019118 UP000030742 UP000283509 UP000092461
UP000053097 UP000027135 UP000007755 UP000242457 UP000078541 UP000005205 UP000078540 UP000007266 UP000000311 UP000008237 UP000235965 UP000078542 UP000279307 UP000002358 UP000053825 UP000215335 UP000005203 UP000076502 UP000053105 UP000019118 UP000030742 UP000283509 UP000092461
PRIDE
Interpro
SUPFAM
SSF48403
SSF48403
Gene 3D
CDD
ProteinModelPortal
H9JYF3
A0A194RKS0
A0A1E1WBA0
A0A3S2LNI4
A0A2W1BKT4
A0A212F6U0
+ More
A0A194Q7H5 A0A2A4IUT9 A0A026W2C9 A0A067R6S1 F4WKQ7 A0A0C9QNM6 A0A2A3E3B3 A0A151JZB1 A0A158NJ94 A0A195BKS7 A0A0C9PMC2 A0A0C9R620 D6WXZ2 E2AXC9 E2BSX3 A0A2J7PJU1 A0A195C3J9 A0A3L8DIY2 A0A1B6H8E1 K7IVV7 A0A0L7R8M7 A0A2J7PJT9 A0A232EE76 A0A1B6FWR0 A0A088A6I0 A0A154PET4 V5GN45 A0A2P6KX80 A0A069DZB8 A0A1B6CRZ0 A0A0P5LKM1 A0A1B6CP52 A0A0M8ZPK8 A0A1B6ECR4 A0A0P5D486 A0A224XLN8 A0A0P5WDV9 A0A0N8CP13 A0A0P6D0J0 A0A0V0GAG1 N6T8M3 A0A023F330 A0A0P5KRH1 A0A0N8D6G2 A0A0P5BGE6 A0A0P5ZEY7 A0A0P5GG21 A0A0P5KYR8 A0A0N8AUK3 A0A0P4ZW21 A0A0P5SHC8 A0A0P5K063 A0A0P5Y1W8 A0A0N8AU28 A0A0N8AV46 A0A0T6AYA9 A0A0P5BF77 A0A0P5U5N9 A0A0N8ATM7 A0A0P4XLU3 A0A0P5WUR3 A0A0P5E596 A0A3R7MMR5 A0A0P6I0I6 A0A0P5D4H6 A0A0P4XKG6 A0A0P4ZVA1 A0A0P5D4R5 A0A0P5VYG8 A0A0N7ZR06 A0A0P5Y1P5 A0A0P5SJD9 A0A0P6DJV7 A0A0P5FLX7 A0A0P6AZP8 A0A0P4Y5N2 A0A0P4Z2X2 A0A0N8E4S1 A0A0P5J336 A0A0N8ADN7 A0A0P5S613 A0A0P5KLE6 A0A1B0GJ11 A0A0P6ACY1 A0A0P5L3A3 A0A1L8DTU1 A0A0N7ZRF1 A0A0P5V8D9 A0A0P4YZ90 A0A0P5JCA0 A0A0P5ZYL2 A0A0P5SKI4
A0A194Q7H5 A0A2A4IUT9 A0A026W2C9 A0A067R6S1 F4WKQ7 A0A0C9QNM6 A0A2A3E3B3 A0A151JZB1 A0A158NJ94 A0A195BKS7 A0A0C9PMC2 A0A0C9R620 D6WXZ2 E2AXC9 E2BSX3 A0A2J7PJU1 A0A195C3J9 A0A3L8DIY2 A0A1B6H8E1 K7IVV7 A0A0L7R8M7 A0A2J7PJT9 A0A232EE76 A0A1B6FWR0 A0A088A6I0 A0A154PET4 V5GN45 A0A2P6KX80 A0A069DZB8 A0A1B6CRZ0 A0A0P5LKM1 A0A1B6CP52 A0A0M8ZPK8 A0A1B6ECR4 A0A0P5D486 A0A224XLN8 A0A0P5WDV9 A0A0N8CP13 A0A0P6D0J0 A0A0V0GAG1 N6T8M3 A0A023F330 A0A0P5KRH1 A0A0N8D6G2 A0A0P5BGE6 A0A0P5ZEY7 A0A0P5GG21 A0A0P5KYR8 A0A0N8AUK3 A0A0P4ZW21 A0A0P5SHC8 A0A0P5K063 A0A0P5Y1W8 A0A0N8AU28 A0A0N8AV46 A0A0T6AYA9 A0A0P5BF77 A0A0P5U5N9 A0A0N8ATM7 A0A0P4XLU3 A0A0P5WUR3 A0A0P5E596 A0A3R7MMR5 A0A0P6I0I6 A0A0P5D4H6 A0A0P4XKG6 A0A0P4ZVA1 A0A0P5D4R5 A0A0P5VYG8 A0A0N7ZR06 A0A0P5Y1P5 A0A0P5SJD9 A0A0P6DJV7 A0A0P5FLX7 A0A0P6AZP8 A0A0P4Y5N2 A0A0P4Z2X2 A0A0N8E4S1 A0A0P5J336 A0A0N8ADN7 A0A0P5S613 A0A0P5KLE6 A0A1B0GJ11 A0A0P6ACY1 A0A0P5L3A3 A0A1L8DTU1 A0A0N7ZRF1 A0A0P5V8D9 A0A0P4YZ90 A0A0P5JCA0 A0A0P5ZYL2 A0A0P5SKI4
Ontologies
GO
PANTHER
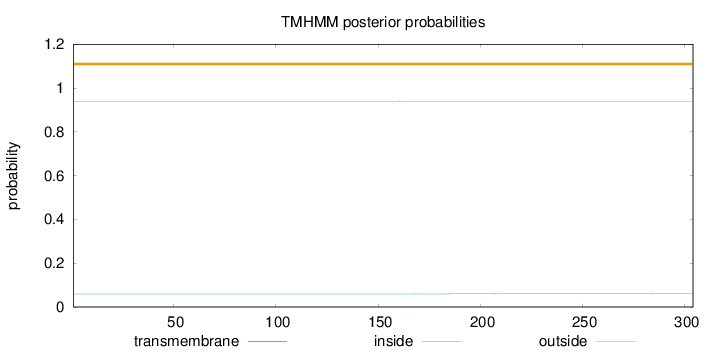
Topology
Length:
304
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01826
Exp number, first 60 AAs:
0.00012
Total prob of N-in:
0.05992
outside
1 - 304
Population Genetic Test Statistics
Pi
250.404364
Theta
151.899825
Tajima's D
2.179942
CLR
0.236265
CSRT
0.910954452277386
Interpretation
Uncertain
