Gene
KWMTBOMO04288
Pre Gene Modal
BGIBMGA012649
Annotation
PREDICTED:_lipase_3-like_[Bombyx_mori]
Full name
Lipase
Location in the cell
PlasmaMembrane Reliability : 2.799
Sequence
CDS
ATGATATTTCCGAGTCTGATTTTTAAAATTGCGTTCATTTTACTCGTCGCCTTTTTAAATAAAGTTTCTAATTCACTTATAGACTCACCGATCTTGGAAAGCATTCCAGGAGACATTGCCATTGAACAAAACGATTTCGTAAAATCGTACGGTTTGGATCCGGTAGGGAATTTAGTGTCCATTGAAGTACCCGAAGACGCTAACCTTGATTTCAACGGTCTCGCCAAGAAGTACGGGCATCCGTGCGAAGAGCATTACATCGAGTCAGAGGATGGTTATATCACCAAGTTATTTCATTTACCGGGAGATAAGAAACGGCCGGTACTCATGATGCACGGCATCTTCGATTCTGCCGATACTTTTATCATCAGAGGCAACAGATCAATGGCCATATCCCTCGCGAACACTGGCTACGACGTGTGGGTGGGAAACATGAGAGGAAACAAACATTCCCGGAGACACGTTACTCTGAATCCCGACAAAGATCCTGAATTTTGGAACTTCAGTTACCACGAAATGGCGTTGTACGATCTACCTGCGACTATCGACTACATCCTGGGTCACACCGGCCAGACACGGATCTATGGTATCGGTCATTCCCTGGGCAACAGTATTTTCTTTGCAATGACGTCAATGTTGCCACAATATAACAAAAAAATCAGGATTTTCATTTCGTTAGCACCGGCTGCTCACCTTGACAATATTAAATTGCCGACAAGAATACTGGCTGTGCTATCTCCCCTTAACAACTATTTGTTGAAAGGTACTGGGCAAGAGGAAATACTTGCGGATTCAACTATCCTGTTTAAAATTTTACGAAAAGTTTGTCCGATTGGTCTGCCTGGATATATTTTATGCGCTGAATTGATTATATTTTACATAGGTGGCTACGATAGGGCAGAACTAGAGCGTGAATTCTTGCCGGTCGTTATAGGTCATGTTACCAGTGGTACCTCAAGGAAAAACTTCATTCACTTGGCGCAGTCGGGTAAATACAAACGTTTCGCACGTTATGATGCCGGAGTCAAAGAAAATCTTCGTGTATACGGAGCCAAGGTACCTCCGGCGTATAATATCAGTGCGGTGACCATGAAAGTAGCTATTTTTGCGGGAAATAACGACAAAATAGTCGTTGTGAAGGACACAAAGAGACTACGAGATGAGTTGAATAATGTAGTGGAATACAGAGTATTGAAACCAAATAAAATTAGCCACTTTGACTTCGTATGGGGAAGGAACGCTAACAGATATTTGTTTCCATTCATATTTGATTTGTTATCGAGTTACTAA
Protein
MIFPSLIFKIAFILLVAFLNKVSNSLIDSPILESIPGDIAIEQNDFVKSYGLDPVGNLVSIEVPEDANLDFNGLAKKYGHPCEEHYIESEDGYITKLFHLPGDKKRPVLMMHGIFDSADTFIIRGNRSMAISLANTGYDVWVGNMRGNKHSRRHVTLNPDKDPEFWNFSYHEMALYDLPATIDYILGHTGQTRIYGIGHSLGNSIFFAMTSMLPQYNKKIRIFISLAPAAHLDNIKLPTRILAVLSPLNNYLLKGTGQEEILADSTILFKILRKVCPIGLPGYILCAELIIFYIGGYDRAELEREFLPVVIGHVTSGTSRKNFIHLAQSGKYKRFARYDAGVKENLRVYGAKVPPAYNISAVTMKVAIFAGNNDKIVVVKDTKRLRDELNNVVEYRVLKPNKISHFDFVWGRNANRYLFPFIFDLLSSY
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Feature
chain Lipase
Uniprot
H9JSY7
H9JT83
H9JYF5
H9JYF4
A0A194Q1A5
A0A0N1IGF0
+ More
A0A0L7L277 A0A1E1WS39 A0A0L7KQV5 A0A2H1VHX0 H9JXK1 A0A194QRY3 A0A2W1BHX8 A0A212F2S9 H9JSY6 I4DP30 A0A194Q1X8 A0A0L7L5K9 A0A212F0W8 H9JSY5 A0A212FGM8 H9JXH3 H9JSX2 A0A212F2S4 A0A2A4JBG8 S4PBS0 A0A2A4J9X8 E2B370 A0A151X254 A0A0L7KXY5 A0A226E1Q5 A0A0T6BEX7 V5I953 A0A1D2MRG2 A0A067R0F9 A0A195FK23 A0A067QXT1 A0A2J7QW32 A0A195E3Z0 A0A0P4WII3 W5J548 B4KKS1 X1WSL9 A0A2J7QRG7 A0A158P269 A0A151IQS0 A0A212F4K7 A0A0C9R2R9 A0A1D2MVM7 A0A1B6HSA1 A0A195AVW7 A0A226EG24 F4W6F9 A0A1B6C389 A0A182H3N2 A0A2A4JCC5 W5JF51 E0V9B6 A0A1B6G3S2 B4LUK3 A0A2A4K633 A0A151X8H0 A0A067QLF8 A0A194R2Y3 A0A0T6BES6 A0A2H1WUD0 A0A1A9UTK2 A0A195EL90 A0A0K8W418 A0A194R3I6 A0A3R7MMP8 A0A2J7QRJ0 A0A195FI51 A0A2W1B637 E2B563 B4KKR3 A0A0L7LE27 B4HWR7 A0A195CI99 A0A067RCF7 A0A0J9R159 A0A2J7QG30 A0A151IQU8 Q16MC6 Q16F27 A0A0K8UT86 A0A2J7QW21 A0A2W1BS78 A0A195FCA6 A0A182RCW3 B0W6G8 W8C916 A0A1B0B8Y7 A0A3L8D6Y6 D2A3G9 F4W6F8 A0A182ML09 A0A151X224 A0A0J9TKN6 E2AR09
A0A0L7L277 A0A1E1WS39 A0A0L7KQV5 A0A2H1VHX0 H9JXK1 A0A194QRY3 A0A2W1BHX8 A0A212F2S9 H9JSY6 I4DP30 A0A194Q1X8 A0A0L7L5K9 A0A212F0W8 H9JSY5 A0A212FGM8 H9JXH3 H9JSX2 A0A212F2S4 A0A2A4JBG8 S4PBS0 A0A2A4J9X8 E2B370 A0A151X254 A0A0L7KXY5 A0A226E1Q5 A0A0T6BEX7 V5I953 A0A1D2MRG2 A0A067R0F9 A0A195FK23 A0A067QXT1 A0A2J7QW32 A0A195E3Z0 A0A0P4WII3 W5J548 B4KKS1 X1WSL9 A0A2J7QRG7 A0A158P269 A0A151IQS0 A0A212F4K7 A0A0C9R2R9 A0A1D2MVM7 A0A1B6HSA1 A0A195AVW7 A0A226EG24 F4W6F9 A0A1B6C389 A0A182H3N2 A0A2A4JCC5 W5JF51 E0V9B6 A0A1B6G3S2 B4LUK3 A0A2A4K633 A0A151X8H0 A0A067QLF8 A0A194R2Y3 A0A0T6BES6 A0A2H1WUD0 A0A1A9UTK2 A0A195EL90 A0A0K8W418 A0A194R3I6 A0A3R7MMP8 A0A2J7QRJ0 A0A195FI51 A0A2W1B637 E2B563 B4KKR3 A0A0L7LE27 B4HWR7 A0A195CI99 A0A067RCF7 A0A0J9R159 A0A2J7QG30 A0A151IQU8 Q16MC6 Q16F27 A0A0K8UT86 A0A2J7QW21 A0A2W1BS78 A0A195FCA6 A0A182RCW3 B0W6G8 W8C916 A0A1B0B8Y7 A0A3L8D6Y6 D2A3G9 F4W6F8 A0A182ML09 A0A151X224 A0A0J9TKN6 E2AR09
Pubmed
EMBL
BABH01038893
BABH01038894
BABH01034815
BABH01043411
KQ459580
KPI99336.1
+ More
KQ460480 KPJ14361.1 JTDY01008561 JTDY01003454 KOB64514.1 KOB69530.1 GDQN01001224 JAT89830.1 JTDY01007194 KOB65389.1 ODYU01002476 SOQ40012.1 BABH01032765 KQ461196 KPJ06301.1 KZ150080 PZC73881.1 AGBW02010690 OWR48036.1 BABH01038896 AK403268 BAM19670.1 KPI99333.1 JTDY01002846 KOB70611.1 AGBW02011015 OWR47367.1 AGBW02008642 OWR52889.1 BABH01032763 BABH01038949 OWR48034.1 NWSH01002215 PCG68894.1 GAIX01002709 JAA89851.1 PCG68895.1 GL445305 EFN89852.1 KQ982580 KYQ54458.1 JTDY01004649 KOB67911.1 LNIX01000009 OXA50396.1 LJIG01001091 KRT85849.1 GALX01003535 JAB64931.1 LJIJ01000666 ODM95481.1 KK852827 KDR15388.1 KQ981523 KYN40354.1 KK853131 KDR10945.1 NEVH01009768 PNF32784.1 KQ979657 KYN19900.1 GDRN01044880 JAI66895.1 ADMH02002130 ETN58533.1 CH933807 EDW12735.1 ABLF02039332 NEVH01011896 PNF31184.1 ADTU01007002 KQ976755 KYN08590.1 AGBW02010351 OWR48653.1 GBYB01007122 JAG76889.1 LJIJ01000475 ODM97066.1 GECU01030185 JAS77521.1 KQ976731 KYM76202.1 LNIX01000004 OXA56097.1 GL887707 EGI70294.1 GEDC01029317 GEDC01024313 GEDC01024096 GEDC01020251 JAS07981.1 JAS12985.1 JAS13202.1 JAS17047.1 JXUM01107700 KQ565165 KXJ71265.1 NWSH01002126 PCG69082.1 ADMH02001587 ETN61958.1 DS234991 EEB09972.1 GECZ01012738 JAS57031.1 CH940649 EDW64189.1 NWSH01000119 PCG79374.1 KQ982409 KYQ56662.1 KK853197 KDR09957.1 KQ460855 KPJ11879.1 LJIG01001092 KRT85848.1 ODYU01011128 SOQ56675.1 KQ978747 KYN28654.1 GDHF01006507 JAI45807.1 KQ460949 KPJ10406.1 QCYY01001042 ROT80984.1 PNF31187.1 KYN40355.1 KZ150291 PZC71579.1 GL445725 EFN89157.1 EDW12727.1 KRG03395.1 JTDY01001496 KOB73727.1 CH480818 EDW52462.1 KQ977720 KYN00465.1 KK852794 KDR16428.1 CM002910 KMY89574.1 NEVH01014836 PNF27554.1 KYN08589.1 CH477866 EAT35491.1 CH478517 EAT32843.1 GDHF01022533 JAI29781.1 PNF32787.1 KZ149971 PZC76037.1 KQ981685 KYN38026.1 DS231848 EDS36642.1 GAMC01007031 JAB99524.1 JXJN01010106 QOIP01000012 RLU16237.1 KQ971338 EFA02315.1 EGI70293.1 AXCM01003911 KYQ54457.1 KMY89575.1 GL441846 EFN64155.1
KQ460480 KPJ14361.1 JTDY01008561 JTDY01003454 KOB64514.1 KOB69530.1 GDQN01001224 JAT89830.1 JTDY01007194 KOB65389.1 ODYU01002476 SOQ40012.1 BABH01032765 KQ461196 KPJ06301.1 KZ150080 PZC73881.1 AGBW02010690 OWR48036.1 BABH01038896 AK403268 BAM19670.1 KPI99333.1 JTDY01002846 KOB70611.1 AGBW02011015 OWR47367.1 AGBW02008642 OWR52889.1 BABH01032763 BABH01038949 OWR48034.1 NWSH01002215 PCG68894.1 GAIX01002709 JAA89851.1 PCG68895.1 GL445305 EFN89852.1 KQ982580 KYQ54458.1 JTDY01004649 KOB67911.1 LNIX01000009 OXA50396.1 LJIG01001091 KRT85849.1 GALX01003535 JAB64931.1 LJIJ01000666 ODM95481.1 KK852827 KDR15388.1 KQ981523 KYN40354.1 KK853131 KDR10945.1 NEVH01009768 PNF32784.1 KQ979657 KYN19900.1 GDRN01044880 JAI66895.1 ADMH02002130 ETN58533.1 CH933807 EDW12735.1 ABLF02039332 NEVH01011896 PNF31184.1 ADTU01007002 KQ976755 KYN08590.1 AGBW02010351 OWR48653.1 GBYB01007122 JAG76889.1 LJIJ01000475 ODM97066.1 GECU01030185 JAS77521.1 KQ976731 KYM76202.1 LNIX01000004 OXA56097.1 GL887707 EGI70294.1 GEDC01029317 GEDC01024313 GEDC01024096 GEDC01020251 JAS07981.1 JAS12985.1 JAS13202.1 JAS17047.1 JXUM01107700 KQ565165 KXJ71265.1 NWSH01002126 PCG69082.1 ADMH02001587 ETN61958.1 DS234991 EEB09972.1 GECZ01012738 JAS57031.1 CH940649 EDW64189.1 NWSH01000119 PCG79374.1 KQ982409 KYQ56662.1 KK853197 KDR09957.1 KQ460855 KPJ11879.1 LJIG01001092 KRT85848.1 ODYU01011128 SOQ56675.1 KQ978747 KYN28654.1 GDHF01006507 JAI45807.1 KQ460949 KPJ10406.1 QCYY01001042 ROT80984.1 PNF31187.1 KYN40355.1 KZ150291 PZC71579.1 GL445725 EFN89157.1 EDW12727.1 KRG03395.1 JTDY01001496 KOB73727.1 CH480818 EDW52462.1 KQ977720 KYN00465.1 KK852794 KDR16428.1 CM002910 KMY89574.1 NEVH01014836 PNF27554.1 KYN08589.1 CH477866 EAT35491.1 CH478517 EAT32843.1 GDHF01022533 JAI29781.1 PNF32787.1 KZ149971 PZC76037.1 KQ981685 KYN38026.1 DS231848 EDS36642.1 GAMC01007031 JAB99524.1 JXJN01010106 QOIP01000012 RLU16237.1 KQ971338 EFA02315.1 EGI70293.1 AXCM01003911 KYQ54457.1 KMY89575.1 GL441846 EFN64155.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000037510
UP000007151
UP000218220
+ More
UP000008237 UP000075809 UP000198287 UP000094527 UP000027135 UP000078541 UP000235965 UP000078492 UP000000673 UP000009192 UP000007819 UP000005205 UP000078542 UP000078540 UP000007755 UP000069940 UP000249989 UP000009046 UP000008792 UP000078200 UP000283509 UP000001292 UP000008820 UP000075900 UP000002320 UP000092460 UP000279307 UP000007266 UP000075883 UP000000311
UP000008237 UP000075809 UP000198287 UP000094527 UP000027135 UP000078541 UP000235965 UP000078492 UP000000673 UP000009192 UP000007819 UP000005205 UP000078542 UP000078540 UP000007755 UP000069940 UP000249989 UP000009046 UP000008792 UP000078200 UP000283509 UP000001292 UP000008820 UP000075900 UP000002320 UP000092460 UP000279307 UP000007266 UP000075883 UP000000311
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9JSY7
H9JT83
H9JYF5
H9JYF4
A0A194Q1A5
A0A0N1IGF0
+ More
A0A0L7L277 A0A1E1WS39 A0A0L7KQV5 A0A2H1VHX0 H9JXK1 A0A194QRY3 A0A2W1BHX8 A0A212F2S9 H9JSY6 I4DP30 A0A194Q1X8 A0A0L7L5K9 A0A212F0W8 H9JSY5 A0A212FGM8 H9JXH3 H9JSX2 A0A212F2S4 A0A2A4JBG8 S4PBS0 A0A2A4J9X8 E2B370 A0A151X254 A0A0L7KXY5 A0A226E1Q5 A0A0T6BEX7 V5I953 A0A1D2MRG2 A0A067R0F9 A0A195FK23 A0A067QXT1 A0A2J7QW32 A0A195E3Z0 A0A0P4WII3 W5J548 B4KKS1 X1WSL9 A0A2J7QRG7 A0A158P269 A0A151IQS0 A0A212F4K7 A0A0C9R2R9 A0A1D2MVM7 A0A1B6HSA1 A0A195AVW7 A0A226EG24 F4W6F9 A0A1B6C389 A0A182H3N2 A0A2A4JCC5 W5JF51 E0V9B6 A0A1B6G3S2 B4LUK3 A0A2A4K633 A0A151X8H0 A0A067QLF8 A0A194R2Y3 A0A0T6BES6 A0A2H1WUD0 A0A1A9UTK2 A0A195EL90 A0A0K8W418 A0A194R3I6 A0A3R7MMP8 A0A2J7QRJ0 A0A195FI51 A0A2W1B637 E2B563 B4KKR3 A0A0L7LE27 B4HWR7 A0A195CI99 A0A067RCF7 A0A0J9R159 A0A2J7QG30 A0A151IQU8 Q16MC6 Q16F27 A0A0K8UT86 A0A2J7QW21 A0A2W1BS78 A0A195FCA6 A0A182RCW3 B0W6G8 W8C916 A0A1B0B8Y7 A0A3L8D6Y6 D2A3G9 F4W6F8 A0A182ML09 A0A151X224 A0A0J9TKN6 E2AR09
A0A0L7L277 A0A1E1WS39 A0A0L7KQV5 A0A2H1VHX0 H9JXK1 A0A194QRY3 A0A2W1BHX8 A0A212F2S9 H9JSY6 I4DP30 A0A194Q1X8 A0A0L7L5K9 A0A212F0W8 H9JSY5 A0A212FGM8 H9JXH3 H9JSX2 A0A212F2S4 A0A2A4JBG8 S4PBS0 A0A2A4J9X8 E2B370 A0A151X254 A0A0L7KXY5 A0A226E1Q5 A0A0T6BEX7 V5I953 A0A1D2MRG2 A0A067R0F9 A0A195FK23 A0A067QXT1 A0A2J7QW32 A0A195E3Z0 A0A0P4WII3 W5J548 B4KKS1 X1WSL9 A0A2J7QRG7 A0A158P269 A0A151IQS0 A0A212F4K7 A0A0C9R2R9 A0A1D2MVM7 A0A1B6HSA1 A0A195AVW7 A0A226EG24 F4W6F9 A0A1B6C389 A0A182H3N2 A0A2A4JCC5 W5JF51 E0V9B6 A0A1B6G3S2 B4LUK3 A0A2A4K633 A0A151X8H0 A0A067QLF8 A0A194R2Y3 A0A0T6BES6 A0A2H1WUD0 A0A1A9UTK2 A0A195EL90 A0A0K8W418 A0A194R3I6 A0A3R7MMP8 A0A2J7QRJ0 A0A195FI51 A0A2W1B637 E2B563 B4KKR3 A0A0L7LE27 B4HWR7 A0A195CI99 A0A067RCF7 A0A0J9R159 A0A2J7QG30 A0A151IQU8 Q16MC6 Q16F27 A0A0K8UT86 A0A2J7QW21 A0A2W1BS78 A0A195FCA6 A0A182RCW3 B0W6G8 W8C916 A0A1B0B8Y7 A0A3L8D6Y6 D2A3G9 F4W6F8 A0A182ML09 A0A151X224 A0A0J9TKN6 E2AR09
PDB
1K8Q
E-value=1.09421e-49,
Score=497
Ontologies
PATHWAY
GO
Topology
SignalP
Position: 1 - 25,
Likelihood: 0.910572
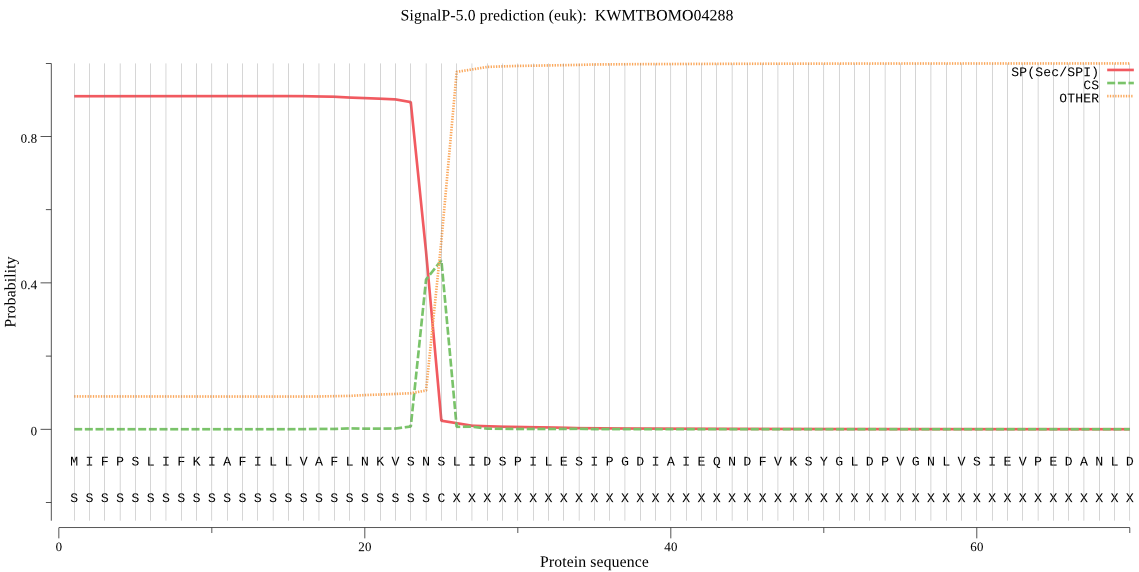
Length:
429
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
14.7135
Exp number, first 60 AAs:
2.14585
Total prob of N-in:
0.11038
outside
1 - 429

Population Genetic Test Statistics
Pi
251.807006
Theta
150.121021
Tajima's D
2.075264
CLR
37.141747
CSRT
0.896555172241388
Interpretation
Uncertain
