Gene
KWMTBOMO04287
Pre Gene Modal
BGIBMGA012648
Annotation
Lipase_[Operophtera_brumata]
Full name
Lipase
Location in the cell
Cytoplasmic Reliability : 1.419 PlasmaMembrane Reliability : 1.728
Sequence
CDS
ATGGTTAAAATTGGCCTTAGGACGTTCTTCATGATATTATCGATCGGCTTACACTTGACATCAGCAGTGTCCATCCTAGAATATCTATTTAATCTAGATGTAAACGAAAACGAGGATTCCAACTTCAAGGACCTGGTCATTAAATACGGTAAGGAATGCGAAGAGTATCAAGTTGTAACGGAGGATGGTTACATTTTAACTCTCTTCCGTATTCCCGGTGACGTCAATCGACCGGTATTGCTTGTGCACGGCTGTTTTGGTTCTGGGGATTCGTTCATCTTAAGGGGTAAGAAGTCGCTGGCTTACGCTCTAGCCGAGGCTGGCTACGACGTGTGGGTCTGCAACTATAGAGGAACTAAATACAGCAGGAAACATGTCACTGTAGATCCAGACCGCGACGACAAGTTCTGGGATTACAGCTTCGAAGAAATCGGTTACTACGACATGCCCGCGTTCATAGACTTCATCTTGGAGAAGACCGGACAAGCCAAGTTACAGACGATAGGATACTCGCAAGGAGGAACGGTCTTCTACGTAATGGGATCTATCAAGCCAGAATACAACGAGAAAATTAAGGTACTCATTTCCCTTGCGCCGATAGCACACTTTTATCACATCAAAAATCCAGTGACCAGGCTGTTCGAAAACGAATCACGTCTGAGTAACATTATCAATGCTATGGGCAAGGAAGAAATTTTTGGTCCACGATCAGGAGAACGAGCTATGATCGACGCGATTTGCTGGAGTCCGTTGACCGGCTATAAGATCTGTTCTCAGTTTTTATTCGAATTCGTTGGTTACGATGAGGACGAATGCGAACCGAAATTCTATTATAAATTAAAGCATTACATACCGAATAGCATTTCGAAAAAGAATGTCCAGCATTTACTCCAATTGAGGCGCGGTTTCAATCGATTCGATTACGGGCCGGAGAGGAACCTTGAGGTCTACAACGCGACATCGCCATCGAAATATGATTTGGATAAGGTCAAGATGAAAATTGTTCTGTACGTATCGAAAAACGATAGGTTGTCAACAGTGGAAGATGGCGAAGAACTACGGAATGAGTTCTCTAACGTTGTTGAATTTCGCATCATCGAACGCGACGAATTCAACCATTTAGATTTTATATGGGCCAAAAATATGGATAAATACATTTTCCCGCACATAATTCGAGCCTTGGAGCAATATAACTTCTAA
Protein
MVKIGLRTFFMILSIGLHLTSAVSILEYLFNLDVNENEDSNFKDLVIKYGKECEEYQVVTEDGYILTLFRIPGDVNRPVLLVHGCFGSGDSFILRGKKSLAYALAEAGYDVWVCNYRGTKYSRKHVTVDPDRDDKFWDYSFEEIGYYDMPAFIDFILEKTGQAKLQTIGYSQGGTVFYVMGSIKPEYNEKIKVLISLAPIAHFYHIKNPVTRLFENESRLSNIINAMGKEEIFGPRSGERAMIDAICWSPLTGYKICSQFLFEFVGYDEDECEPKFYYKLKHYIPNSISKKNVQHLLQLRRGFNRFDYGPERNLEVYNATSPSKYDLDKVKMKIVLYVSKNDRLSTVEDGEELRNEFSNVVEFRIIERDEFNHLDFIWAKNMDKYIFPHIIRALEQYNF
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Feature
chain Lipase
Uniprot
H9JSY6
A0A0L7L277
H9JXK1
A0A0N1IGF0
A0A194Q1A5
A0A0L7KQV5
+ More
A0A2H1VHX0 A0A212F2S9 H9JYF4 H9JT83 H9JXH3 H9JSY7 A0A2W1BHX8 A0A1E1WS39 H9JYF5 A0A194QRY3 H9JSX2 I4DP30 A0A0L7L5K9 A0A194Q1X8 A0A212F0W8 A0A212F2S4 S4PBS0 A0A212FGM8 H9JSY5 V5I953 B4LUL0 A0A2A4JBG8 A0A0Q9WAE5 A0A2A4J9X8 B4JE90 B4JE91 A0A0M5J114 B4KKS2 A0A0J9TKN6 A0A3B0KDQ2 A0A194R3I6 B4HWR7 A0A067QLF8 A0A0J9R159 A0A0L7LE27 A0A0L7KXY5 M9MRM3 A0A3B0JHG8 A0A0A9XKD9 A0A146LJT0 A0A0P8XGJ3 W5JF51 A0A1W7R7P1 A0A2J7QW32 A0A1W4UT82 A0A182RXX8 A0A0Q5WBH0 A0A0R3NZX2 Q9VKT9 B3MJK2 A0A0K8SGC1 A0A0R1DRH5 A0A182GFS9 M9PCU5 D2A0X3 A0A2A4K633 E2AR09 A0A194PMH9 A0A1W4UT02 A0A0Q9WQJ1 A0A0K8W418 A0A1S4F923 A0A182RCW3 A0A182VQ94 E2B370 B3N5D9 B3MJK3 Q29KR9 A0A182RXY7 A0A067R0F9 B4P0Q9 A0A034VW56 A0A182FR70 A0A067RCF7 B4GSW4 A0A2A4JCC5 A0A2W1B637 Q17BM4 B4MWR9 A0A182H3N4 B4Q9C6 A0A2H1WJP8 A0A182MKD5 A0A1D2N240 A0A182H3N2 Q16F25 Q16MC9 A0A212FGL9 A0A194PRT8 A0A088A7F6 A0A0J9R0A7 A0A026X0Q1 W8C916 A0A1S4FWC2
A0A2H1VHX0 A0A212F2S9 H9JYF4 H9JT83 H9JXH3 H9JSY7 A0A2W1BHX8 A0A1E1WS39 H9JYF5 A0A194QRY3 H9JSX2 I4DP30 A0A0L7L5K9 A0A194Q1X8 A0A212F0W8 A0A212F2S4 S4PBS0 A0A212FGM8 H9JSY5 V5I953 B4LUL0 A0A2A4JBG8 A0A0Q9WAE5 A0A2A4J9X8 B4JE90 B4JE91 A0A0M5J114 B4KKS2 A0A0J9TKN6 A0A3B0KDQ2 A0A194R3I6 B4HWR7 A0A067QLF8 A0A0J9R159 A0A0L7LE27 A0A0L7KXY5 M9MRM3 A0A3B0JHG8 A0A0A9XKD9 A0A146LJT0 A0A0P8XGJ3 W5JF51 A0A1W7R7P1 A0A2J7QW32 A0A1W4UT82 A0A182RXX8 A0A0Q5WBH0 A0A0R3NZX2 Q9VKT9 B3MJK2 A0A0K8SGC1 A0A0R1DRH5 A0A182GFS9 M9PCU5 D2A0X3 A0A2A4K633 E2AR09 A0A194PMH9 A0A1W4UT02 A0A0Q9WQJ1 A0A0K8W418 A0A1S4F923 A0A182RCW3 A0A182VQ94 E2B370 B3N5D9 B3MJK3 Q29KR9 A0A182RXY7 A0A067R0F9 B4P0Q9 A0A034VW56 A0A182FR70 A0A067RCF7 B4GSW4 A0A2A4JCC5 A0A2W1B637 Q17BM4 B4MWR9 A0A182H3N4 B4Q9C6 A0A2H1WJP8 A0A182MKD5 A0A1D2N240 A0A182H3N2 Q16F25 Q16MC9 A0A212FGL9 A0A194PRT8 A0A088A7F6 A0A0J9R0A7 A0A026X0Q1 W8C916 A0A1S4FWC2
Pubmed
19121390
26227816
26354079
22118469
28756777
22651552
+ More
23622113 17994087 22936249 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25401762 26823975 20920257 23761445 15632085 26109357 26109356 17550304 26483478 18362917 19820115 20798317 25348373 17510324 27289101 24508170 24495485
23622113 17994087 22936249 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25401762 26823975 20920257 23761445 15632085 26109357 26109356 17550304 26483478 18362917 19820115 20798317 25348373 17510324 27289101 24508170 24495485
EMBL
BABH01038896
JTDY01008561
JTDY01003454
KOB64514.1
KOB69530.1
BABH01032765
+ More
KQ460480 KPJ14361.1 KQ459580 KPI99336.1 JTDY01007194 KOB65389.1 ODYU01002476 SOQ40012.1 AGBW02010690 OWR48036.1 BABH01043411 BABH01034815 BABH01032763 BABH01038893 BABH01038894 KZ150080 PZC73881.1 GDQN01001224 JAT89830.1 KQ461196 KPJ06301.1 BABH01038949 AK403268 BAM19670.1 JTDY01002846 KOB70611.1 KPI99333.1 AGBW02011015 OWR47367.1 OWR48034.1 GAIX01002709 JAA89851.1 AGBW02008642 OWR52889.1 GALX01003535 JAB64931.1 CH940649 EDW64196.1 NWSH01002215 PCG68894.1 KRF81490.1 PCG68895.1 CH916368 EDW03610.1 EDW03611.1 CP012523 ALC38094.1 CH933807 EDW12736.1 CM002910 KMY89575.1 OUUW01000006 SPP81788.1 KQ460949 KPJ10406.1 CH480818 EDW52462.1 KK853197 KDR09957.1 KMY89574.1 JTDY01001496 KOB73727.1 JTDY01004649 KOB67911.1 AE014134 ADV37035.1 SPP81787.1 GBHO01027632 GBHO01024326 GBHO01024325 JAG15972.1 JAG19278.1 JAG19279.1 GDHC01011084 JAQ07545.1 CH902620 KPU73921.1 ADMH02001587 ETN61958.1 GEHC01000530 JAV47115.1 NEVH01009768 PNF32784.1 CH954177 KQS70596.1 CH379061 KRT04741.1 AY089586 AAF52971.1 AAL90324.1 EDV32370.2 GBRD01013485 JAG52341.1 CM000157 KRJ97507.1 JXUM01060253 KQ562094 KXJ76706.1 AGB92891.1 KQ971338 EFA01616.1 NWSH01000119 PCG79374.1 GL441846 EFN64155.1 KQ459598 KPI94527.1 CH963857 KRF98484.1 GDHF01006507 JAI45807.1 GL445305 EFN89852.1 EDV59018.1 EDV32371.1 EAL33105.2 KK852827 KDR15388.1 EDW87954.1 GAKP01013224 JAC45728.1 KK852794 KDR16428.1 CH479189 EDW25473.1 NWSH01002126 PCG69082.1 KZ150291 PZC71579.1 CH477320 EAT43642.1 EDW76558.1 JXUM01107700 KQ565165 KXJ71267.1 CM000361 EDX04568.1 ODYU01009113 SOQ53300.1 AXCM01001786 LJIJ01000287 ODM99328.1 KXJ71265.1 CH478517 EAT32845.1 CH477866 EAT35489.1 AGBW02008645 OWR52878.1 KQ459596 KPI95464.1 KMY89576.1 KK107054 EZA61581.1 GAMC01007031 JAB99524.1
KQ460480 KPJ14361.1 KQ459580 KPI99336.1 JTDY01007194 KOB65389.1 ODYU01002476 SOQ40012.1 AGBW02010690 OWR48036.1 BABH01043411 BABH01034815 BABH01032763 BABH01038893 BABH01038894 KZ150080 PZC73881.1 GDQN01001224 JAT89830.1 KQ461196 KPJ06301.1 BABH01038949 AK403268 BAM19670.1 JTDY01002846 KOB70611.1 KPI99333.1 AGBW02011015 OWR47367.1 OWR48034.1 GAIX01002709 JAA89851.1 AGBW02008642 OWR52889.1 GALX01003535 JAB64931.1 CH940649 EDW64196.1 NWSH01002215 PCG68894.1 KRF81490.1 PCG68895.1 CH916368 EDW03610.1 EDW03611.1 CP012523 ALC38094.1 CH933807 EDW12736.1 CM002910 KMY89575.1 OUUW01000006 SPP81788.1 KQ460949 KPJ10406.1 CH480818 EDW52462.1 KK853197 KDR09957.1 KMY89574.1 JTDY01001496 KOB73727.1 JTDY01004649 KOB67911.1 AE014134 ADV37035.1 SPP81787.1 GBHO01027632 GBHO01024326 GBHO01024325 JAG15972.1 JAG19278.1 JAG19279.1 GDHC01011084 JAQ07545.1 CH902620 KPU73921.1 ADMH02001587 ETN61958.1 GEHC01000530 JAV47115.1 NEVH01009768 PNF32784.1 CH954177 KQS70596.1 CH379061 KRT04741.1 AY089586 AAF52971.1 AAL90324.1 EDV32370.2 GBRD01013485 JAG52341.1 CM000157 KRJ97507.1 JXUM01060253 KQ562094 KXJ76706.1 AGB92891.1 KQ971338 EFA01616.1 NWSH01000119 PCG79374.1 GL441846 EFN64155.1 KQ459598 KPI94527.1 CH963857 KRF98484.1 GDHF01006507 JAI45807.1 GL445305 EFN89852.1 EDV59018.1 EDV32371.1 EAL33105.2 KK852827 KDR15388.1 EDW87954.1 GAKP01013224 JAC45728.1 KK852794 KDR16428.1 CH479189 EDW25473.1 NWSH01002126 PCG69082.1 KZ150291 PZC71579.1 CH477320 EAT43642.1 EDW76558.1 JXUM01107700 KQ565165 KXJ71267.1 CM000361 EDX04568.1 ODYU01009113 SOQ53300.1 AXCM01001786 LJIJ01000287 ODM99328.1 KXJ71265.1 CH478517 EAT32845.1 CH477866 EAT35489.1 AGBW02008645 OWR52878.1 KQ459596 KPI95464.1 KMY89576.1 KK107054 EZA61581.1 GAMC01007031 JAB99524.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000053268
UP000007151
UP000008792
+ More
UP000218220 UP000001070 UP000092553 UP000009192 UP000268350 UP000001292 UP000027135 UP000000803 UP000007801 UP000000673 UP000235965 UP000192221 UP000075900 UP000008711 UP000001819 UP000002282 UP000069940 UP000249989 UP000007266 UP000000311 UP000007798 UP000075920 UP000008237 UP000069272 UP000008744 UP000008820 UP000000304 UP000075883 UP000094527 UP000005203 UP000053097
UP000218220 UP000001070 UP000092553 UP000009192 UP000268350 UP000001292 UP000027135 UP000000803 UP000007801 UP000000673 UP000235965 UP000192221 UP000075900 UP000008711 UP000001819 UP000002282 UP000069940 UP000249989 UP000007266 UP000000311 UP000007798 UP000075920 UP000008237 UP000069272 UP000008744 UP000008820 UP000000304 UP000075883 UP000094527 UP000005203 UP000053097
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9JSY6
A0A0L7L277
H9JXK1
A0A0N1IGF0
A0A194Q1A5
A0A0L7KQV5
+ More
A0A2H1VHX0 A0A212F2S9 H9JYF4 H9JT83 H9JXH3 H9JSY7 A0A2W1BHX8 A0A1E1WS39 H9JYF5 A0A194QRY3 H9JSX2 I4DP30 A0A0L7L5K9 A0A194Q1X8 A0A212F0W8 A0A212F2S4 S4PBS0 A0A212FGM8 H9JSY5 V5I953 B4LUL0 A0A2A4JBG8 A0A0Q9WAE5 A0A2A4J9X8 B4JE90 B4JE91 A0A0M5J114 B4KKS2 A0A0J9TKN6 A0A3B0KDQ2 A0A194R3I6 B4HWR7 A0A067QLF8 A0A0J9R159 A0A0L7LE27 A0A0L7KXY5 M9MRM3 A0A3B0JHG8 A0A0A9XKD9 A0A146LJT0 A0A0P8XGJ3 W5JF51 A0A1W7R7P1 A0A2J7QW32 A0A1W4UT82 A0A182RXX8 A0A0Q5WBH0 A0A0R3NZX2 Q9VKT9 B3MJK2 A0A0K8SGC1 A0A0R1DRH5 A0A182GFS9 M9PCU5 D2A0X3 A0A2A4K633 E2AR09 A0A194PMH9 A0A1W4UT02 A0A0Q9WQJ1 A0A0K8W418 A0A1S4F923 A0A182RCW3 A0A182VQ94 E2B370 B3N5D9 B3MJK3 Q29KR9 A0A182RXY7 A0A067R0F9 B4P0Q9 A0A034VW56 A0A182FR70 A0A067RCF7 B4GSW4 A0A2A4JCC5 A0A2W1B637 Q17BM4 B4MWR9 A0A182H3N4 B4Q9C6 A0A2H1WJP8 A0A182MKD5 A0A1D2N240 A0A182H3N2 Q16F25 Q16MC9 A0A212FGL9 A0A194PRT8 A0A088A7F6 A0A0J9R0A7 A0A026X0Q1 W8C916 A0A1S4FWC2
A0A2H1VHX0 A0A212F2S9 H9JYF4 H9JT83 H9JXH3 H9JSY7 A0A2W1BHX8 A0A1E1WS39 H9JYF5 A0A194QRY3 H9JSX2 I4DP30 A0A0L7L5K9 A0A194Q1X8 A0A212F0W8 A0A212F2S4 S4PBS0 A0A212FGM8 H9JSY5 V5I953 B4LUL0 A0A2A4JBG8 A0A0Q9WAE5 A0A2A4J9X8 B4JE90 B4JE91 A0A0M5J114 B4KKS2 A0A0J9TKN6 A0A3B0KDQ2 A0A194R3I6 B4HWR7 A0A067QLF8 A0A0J9R159 A0A0L7LE27 A0A0L7KXY5 M9MRM3 A0A3B0JHG8 A0A0A9XKD9 A0A146LJT0 A0A0P8XGJ3 W5JF51 A0A1W7R7P1 A0A2J7QW32 A0A1W4UT82 A0A182RXX8 A0A0Q5WBH0 A0A0R3NZX2 Q9VKT9 B3MJK2 A0A0K8SGC1 A0A0R1DRH5 A0A182GFS9 M9PCU5 D2A0X3 A0A2A4K633 E2AR09 A0A194PMH9 A0A1W4UT02 A0A0Q9WQJ1 A0A0K8W418 A0A1S4F923 A0A182RCW3 A0A182VQ94 E2B370 B3N5D9 B3MJK3 Q29KR9 A0A182RXY7 A0A067R0F9 B4P0Q9 A0A034VW56 A0A182FR70 A0A067RCF7 B4GSW4 A0A2A4JCC5 A0A2W1B637 Q17BM4 B4MWR9 A0A182H3N4 B4Q9C6 A0A2H1WJP8 A0A182MKD5 A0A1D2N240 A0A182H3N2 Q16F25 Q16MC9 A0A212FGL9 A0A194PRT8 A0A088A7F6 A0A0J9R0A7 A0A026X0Q1 W8C916 A0A1S4FWC2
PDB
1K8Q
E-value=1.68103e-47,
Score=478
Ontologies
GO
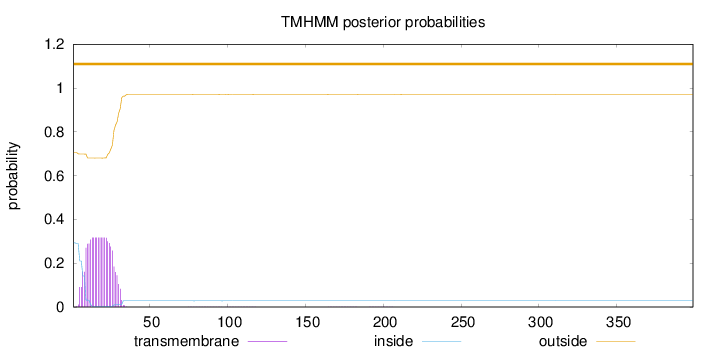
Topology
SignalP
Position: 1 - 22,
Likelihood: 0.894763

Length:
399
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.73380999999999
Exp number, first 60 AAs:
6.6911
Total prob of N-in:
0.29386
outside
1 - 399
Population Genetic Test Statistics
Pi
425.771937
Theta
194.139198
Tajima's D
3.741945
CLR
0.299762
CSRT
0.996800159992
Interpretation
Uncertain
