Gene
KWMTBOMO04283 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012651
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_LOC105841899?_partial_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.223
Sequence
CDS
ATGCAAACTTGTCTTAAAAATGTCATTAGCGAGATTCAAGTGTCCGTCAAATATTTTGTGTTCTGTGATGGCTACCGCCACACTCTGGTCAGTTCAGGCAACGGTTGGCTCGCCAAGCCTCCCCTTTGGGGGCTCACCCGATATCTGTCAAGACTATTCGAAAGTATGTACGCGCCGCCGGAATACGACGGTGAAGTCCCTGCTGAGGTGCCCCATATAACGGGGGCGGAGCTCCGTGTGGCCGTGCGCAAAATGTGTGCGAAGGACACTGCACCCGGCCCGGACGGCGTCCATGGCCGGGTTTGGGCCTTGGCTCTTGGTGCCCTAGGGGACCGACTCTTGAGGCTTTACAACTCCTGCCTCGAGTCGGGACGGTTTCCTTCATCGTGGAGGACGGGCAGACTCGTGCTGTTGAGAAAGGAAGGGCGCCCGGCCGATACTGCCGCCGGGTACCGTCCCATCGTGTTGCTGGATGAGGTGGGCAAGCTGCTGGAACGCATTCTGGCGGCCCGCATCATCCAGCATCTGGTCAGGGTGGGACCCGATCTGTCGGCGGAGCAGTATGGCTTCCGAGAGGGCCGCTCAACGGTAGACGCGATCCTTCGCGTGCGGGCCCTCTCGGAGGAGGCCGTCTCCAGGGGTGGGGTGGCTTTGGCGGTGTCGCTCGACATCGCCAATGCGTTTAACACCCTGCCCTGGTCCGTGATAAGGGGGGCGCTGGAACGACATGGAGTGCCTCCCTACCTTCGCCGGCTGGTGGGTTCCTACTTGGAGGACAGATCGGTCACGTGTACCGGACACGGTGGGATCCTGCACCGGTTCCCGGTCGTGCGCGGTGTTCCACAGGGGTCGGTGCTCGGCCCCCTTTTGTGGAATATCGGGTATGACTGGGTGCTGAGAGGTGCCCTCCTCCCGGGCCTGAACGTAATCTGTTACGAAGACGACACGTTGGTCGTGGCCCGGGGGGGTAGTTTTGCCGAGTCTGCCCGTCTTGCTACCGCTGGGGTGGCGCATGTCGTCGGCAAAATCAGGAGATTGGGCCTCGACGTGGCGCTCAGTAAATCCGAGGCCATGTGGTTCCACAGGCCCCGGAGAGTGCCACCTGTCGATGCCCATATCGTGAGCCTGGTCCCTCGTTTGTTGGGGGTGGCCGGCGCGTTAAGCCGGCTTCTGCCCAACGTCGGGGGGCCTGACCAGGTGACGCGCCGTCTCTATACGGGGGTGGTGCGATCAATGGCCCTATACGGGGCACCTGTGTGGGGCCAGTCCCTGGCCGCGGGGGTGGCGAAGCTGCTGCAACGGCCGCAACGCACCATCGCGGTCAGGGTCATCCGTGGTTATCGCACCATCTCCTTTGAGGCGGCGTGTGTACTGGCTGGGACGCCGCCTTGGGTTCTGGAAGCGGAGGCGCTCGCCGCTGACTATCAGTGGCGGGCTGACCTTCGTGCCCGGGACGTGGCGCGTCCCAGCCCCAGTGTGGTCAGAGCGCGGAGGGCCCAATCTCGGCGACGGCTGGCTGATCCTTCGGCTGGTCGTAGGACCGTCGAGGCGATTCGCCCGGTCCTTGTGGATTGGGTGAATCGTGACAGAGGACGCCTCACTTTCCGGCTCACGCAGGTGCTCACTGGGCATGGTTGCTTCGGTGAGTTCCTGCACCGGATCACAGCCGAGCCGACGGCAGAGTGCCACCATTGTGGTTGCGACTTCGACACGGCAGAGCATACGCTCGTCGCCTGCCCCGCATGGGAGGGGTGGCGCCGTGTCCTCGTCGCAAAAATAGGAAACGACTTGTCGTTGCCGAGTGTTGTGGCATCGATGCTCGGCGACGACGAGTCGTGGAAGGCGATGCTCGACTTCTGCGAGTGCACCATCTCGCAGAAGGAGGCGGCGGGGCGCGCCACGACGACTTCACGTGGGGGTGCGTTCTGGTGGTCGCCCGAGATCGCGAGACTCCGTGAGGAGTGCGTGAGGGCGCGCCGCCGGAGCGCACGCCACCGCCGCCGCCGTCGTCGCGACGCTGCGTTCGCGGAGACGGCGGCCCAGCTGCACGCTGACTGTCGTCAAAAGCAGACGGCGCTGCAGCTGGCCATCGGCGAGGCCAAGAAGCAGAGCATGAAGACTCTCCTGGAGTCGCTCGACGAAGATCCCTGGGGGCGCCCATACAAGATGGTTCGCAGGAAACTGCAGCCGTGGGCGCTCCCGGTGACCGAGCGGCTCCAGCCTCGGCAGCTGCGGGAGATAGTTTCCGCGCTTTTCCCGCCGGCAGCGGGGGGGGACTTTGAGCCCCCCCAGATGGACGCCACGTGGGGCACACCTCCACGTCGCCCCAGCGTTCCCGCTGCCGAGGAGCCCCCTCCCCCTATCACGGGGGCGGAGATCCATGCGGCCGTGTCCAAAATGAGAGCGAAGGACACGGCCCCCGGCCCTGACGGCGTTCACGGCCGGGTCTGGAGCTTGGCCTTCGAGGCCCTAGGGGACCGGCTCGGGGGGCTTTTCGAAGCGTGCCTCGAGTCGGGACGGTTCCCGTCGAAATGGAAGACGGGCAGACTGGTCCTTCTGCGGAAGGACGGGCGCCCCGCGGACTCACCCGCGGGCTATCGCCCCATCGTGTTGCTGGACGAAGCGGGCAAGATGCTCGAGAGAATCGTCGCCGCCCGCATCGTCCGGCACCTGACCGAGACGGGTCCCGATCTTTCGGCGGAGCAATACGGCTTCAGAGAGGGCCGCTCGACTATCGACGCAATTCTGCGCGTGCGGGCCCTCTCCGACGAAGCCGTATCCCGGGGCGGGGTGGCGATGGCGCTATCCTTAGATGTAAGGAACGCCTTCAACACCCTGCCCTGGTCGGTGATCGCGGGGGCGCTGGAGTATCACGGCGTCCCCGCATACCTCCGCCGACTGATCGGCTCCTACCTCGAGGGCAGGTCGATCCAGTGCATCGGACACGGTGGGACGATGTACCGCTTCCCCGTCGAGCGCGGTGTTCCGCAGGGGTCTGTCCTGGGCCCCCTGCTGTGGAACATCGGCTACGACTGGGTCCTGCGGGGCGTCATTCGGGGTCCCCTCCCCGGGCTGAGCGTAATATGTTACGCCGACGACACGTTGGTCGTGGCTCCGGGGAGGGACTACCGGGAGTCTGCCCGTCTGGCGTGCGCAGGCGTGGCACACGTCGTCACTAGGATCCGACGGTTGGGGCTCGAGGTGGCGCTCGATAAAACCCAGGCCCTGCTTTTTCACGGGCCGGGACGAGCGCCGCCTGTGGGTGCCCACCTCGTGATCGGAGGCGTCCGCGTCGGGGTTGGGGTGACCGGTCTTCGGTACCTCGGCCTCGAACTGGATAGTCGGTGGAACTTCCGCGCTCACTTTGAGAAGTTGGGCCCTCGACTGATGGCGACGGCCGGCTCTCTGAGCCGGCTGCTTCCAAACGTCGGGGGTCCCGATCAGGTGGCGCGCCGCCTCTACATGGGGGTGGTGCGGTCGATGGCACTATACGGGGCTCCCGTATGGTGCCGCGCCCTGACCCGCAAGAACGTCGCGGCTCTGCGACGTCCGCAGCGTGCGATCGCGGTCAGGGCGATCCGAGGATACCGCACCGTCTCCTTCGAGGCGGCGTGCTTGCTCGCCGGGGCGCCACCCTGGGACCTGGAGGCGGAGGCGCTCGCTGCCGATTACAGGTGGCGTAGCGACCTCCGCTCTAGGGGGGAAGGGCGCCCCGGCGAAGGTGTAGTTCGAGCGCGGAGGCTCCACTCTCGGCGGTCCGTGCTGGAGGCGTGGTCTCGCCGCTTGGCGGACCCGTCGGCCGGCCTCCGAACCGTCGAGGCGGTCCGCCCGGTTCTCGCGGACTGGGTGGGCCGCGACCGCGGATGCCTCACCTTCCGGCTAACGCAGATGCTTACCGGCCATGGTTGTTTCGGCCGGTACTTGCACAAGATAGCCGGAAGGGAACCGACGGCGCAGTGCCACCACTGCGCGGACCGCGACGAGGAAGACACAGCGGAACACACGCTGGCGCGTTGCTCTGGATTCGACGAGCAACGCGCCGCCCTCGTCGCGGTCATAGGAGAGGACCTCTCGCTGCCGCGCGTCGTGGCTACGATGCTCGGCAGCGACGCGTCCTGGAAGGCGATGCTCGACTTCTGCGAGTCCACCATCTCGCAGAAGGAGGCGGCGGAGCGAGAGAGGGAGAGCTCTTCCCTTTCCGCACCGATCCGTCGCCGTCGAGCCGGGGGCCGGAGGCGGGAATACGCCCATAATCTCAACCCAACTGGGATGTCTTTGAAGAATAGGTTAATTGCATGTCTGCAAAAACTGAAGGAATACGATTACACTGACGTAGACAAGGCAAGAGCTAGTGACGCGAAGGCCGAGAATGCCCTCAACTTAATATACCAGACTGCTTTAAAGAAAACAACTTTTACACCAGAAGAGAAGAAAATTGTAGGTTCTCTTTTATTTGATGCCATCACACCGATACAAAGCAACATAGAAGGAACGCTCTGCGAATACAGAACTAGATTAGATACCTCTGTGCTTATGAAAAGATCTGCAATACAATTCTTGATTGACGAATACGGGAAGTTTCCAGTTGAGAATTCTATCTTGGCTACAAAGTTAGAGGAAGCCGGCATAGCAGAGAGCGTTGAAATACTCAATGAAATTATAAATAGGACATCGGCGGTCCTAGCAGGAGGAGCAGTTGGATGCCTCGCAGGACCGGCAGGAGCTGCATTGGGAGGCGTATACGGAGGCATGATTACAGACGGCTTGGTTTCTGCGGCTAAGAAGGAGCCTGTAGGTATTTTTGAGTCAAGTGCAAATATAGGTAGAGACATTGCTTCAGGCAAAAATCCAATGGCTTCTGTGGGTTCCGCAAGTTTCAAAATCGCCATGGACGGTATAGGTGGCTTTGGCATGGGAGGAGTTTCTAGCGTTGCCAGTAAAGTCCCGGCGAAGGCCGTCGCAAAAACCATAGAAGGCACTGTAAAAAAAGCTGCTGCAACTATTGGGGAGGAGATGATTGAGAAGTCAGTGCAATTAACTGCAGAAGAAATGACAAAAATTGTGGCAAAGAAAGTGTCAGAAAAAGCCGTGAAAAAAGCTGTCGAAACTGCAGCAATACAAACACAAATGCTTCTGGTGTCTGCTCACGTCGGAGTAAATGCTAGTGAAAAAACCAAAGCTGAAATGCGTGCTGAAGAAGAACGTAAACAACAGAAACAAAAAGAAGAAGAAAAAAAGGCGCGTGAAAAGCGCGATAAAAAAAACACTGGATCGTGGGAACCACCACGCGAGAACAGAAACAATAAAGAGTCCAATGTTGATAATGAAGAGTCCGACGAGGAGGTATTTGAAAAGTTCATGAAATTACTTAAACAGCTCTTGAAGCTTCTATACGGCAATAACAGCGAACCGTTCACAAGAATTTTCGAGAATTTATCACCAGGCCAAATAGCGTATTTGGTGAAAATAATCAGAAACCTTAACTCAACTGGAATTTTACAGATAGTGTTAGATTTTACATTAAAACTGTTTAAATTGTACTTTTCTAAAGATATCTCATTCACAAATATCCTGAGTATGTTAGAATTCGTCGCAACTTACATTGAAGACCAGTTGTATTTTATGACCAAAACCAAAGGTAAACCGACTCGTCAATCAGATGCCGACCGGAATGAGCGTCGAAGGAACGCCATCAGAGAACTTCAAAACCAAAGGGATATAATTCAGAAAATTGCTGATCTATTCAGACATTTACAAGCGGAAATCATAGCTGAAACGAGCGCGCCTAATATAATTTCCCCCTATTTCCGTGATATATTCTCAGCTGTGTATCATTATTACAAACACAGACGCGTGCCCGTGCCAGGCCATGAGAAGCTGACAGTCAAACAATATTTCGACTTGATAAGGATGGTAATTATAAAAATGAAAGAATCACCGGAATACAAAAAAATGCAGAAGGCCAGAAATGGAACGACCCTCACTTGTGAAATGTACGTGGTCTTGTTCGGTAGGATGTTCAGAATTATTATTACTGTTCGAGATGGTGAAATTGTATTGAGTACCGCCTATGTAGTCAAGCGTATGAACTTTAATGCCAAAGAAGGCATTCTTTACATAAAATTCGGTGATGGAGATGTACAGAAGTAA
Protein
MQTCLKNVISEIQVSVKYFVFCDGYRHTLVSSGNGWLAKPPLWGLTRYLSRLFESMYAPPEYDGEVPAEVPHITGAELRVAVRKMCAKDTAPGPDGVHGRVWALALGALGDRLLRLYNSCLESGRFPSSWRTGRLVLLRKEGRPADTAAGYRPIVLLDEVGKLLERILAARIIQHLVRVGPDLSAEQYGFREGRSTVDAILRVRALSEEAVSRGGVALAVSLDIANAFNTLPWSVIRGALERHGVPPYLRRLVGSYLEDRSVTCTGHGGILHRFPVVRGVPQGSVLGPLLWNIGYDWVLRGALLPGLNVICYEDDTLVVARGGSFAESARLATAGVAHVVGKIRRLGLDVALSKSEAMWFHRPRRVPPVDAHIVSLVPRLLGVAGALSRLLPNVGGPDQVTRRLYTGVVRSMALYGAPVWGQSLAAGVAKLLQRPQRTIAVRVIRGYRTISFEAACVLAGTPPWVLEAEALAADYQWRADLRARDVARPSPSVVRARRAQSRRRLADPSAGRRTVEAIRPVLVDWVNRDRGRLTFRLTQVLTGHGCFGEFLHRITAEPTAECHHCGCDFDTAEHTLVACPAWEGWRRVLVAKIGNDLSLPSVVASMLGDDESWKAMLDFCECTISQKEAAGRATTTSRGGAFWWSPEIARLREECVRARRRSARHRRRRRRDAAFAETAAQLHADCRQKQTALQLAIGEAKKQSMKTLLESLDEDPWGRPYKMVRRKLQPWALPVTERLQPRQLREIVSALFPPAAGGDFEPPQMDATWGTPPRRPSVPAAEEPPPPITGAEIHAAVSKMRAKDTAPGPDGVHGRVWSLAFEALGDRLGGLFEACLESGRFPSKWKTGRLVLLRKDGRPADSPAGYRPIVLLDEAGKMLERIVAARIVRHLTETGPDLSAEQYGFREGRSTIDAILRVRALSDEAVSRGGVAMALSLDVRNAFNTLPWSVIAGALEYHGVPAYLRRLIGSYLEGRSIQCIGHGGTMYRFPVERGVPQGSVLGPLLWNIGYDWVLRGVIRGPLPGLSVICYADDTLVVAPGRDYRESARLACAGVAHVVTRIRRLGLEVALDKTQALLFHGPGRAPPVGAHLVIGGVRVGVGVTGLRYLGLELDSRWNFRAHFEKLGPRLMATAGSLSRLLPNVGGPDQVARRLYMGVVRSMALYGAPVWCRALTRKNVAALRRPQRAIAVRAIRGYRTVSFEAACLLAGAPPWDLEAEALAADYRWRSDLRSRGEGRPGEGVVRARRLHSRRSVLEAWSRRLADPSAGLRTVEAVRPVLADWVGRDRGCLTFRLTQMLTGHGCFGRYLHKIAGREPTAQCHHCADRDEEDTAEHTLARCSGFDEQRAALVAVIGEDLSLPRVVATMLGSDASWKAMLDFCESTISQKEAAERERESSSLSAPIRRRRAGGRRREYAHNLNPTGMSLKNRLIACLQKLKEYDYTDVDKARASDAKAENALNLIYQTALKKTTFTPEEKKIVGSLLFDAITPIQSNIEGTLCEYRTRLDTSVLMKRSAIQFLIDEYGKFPVENSILATKLEEAGIAESVEILNEIINRTSAVLAGGAVGCLAGPAGAALGGVYGGMITDGLVSAAKKEPVGIFESSANIGRDIASGKNPMASVGSASFKIAMDGIGGFGMGGVSSVASKVPAKAVAKTIEGTVKKAAATIGEEMIEKSVQLTAEEMTKIVAKKVSEKAVKKAVETAAIQTQMLLVSAHVGVNASEKTKAEMRAEEERKQQKQKEEEKKAREKRDKKNTGSWEPPRENRNNKESNVDNEESDEEVFEKFMKLLKQLLKLLYGNNSEPFTRIFENLSPGQIAYLVKIIRNLNSTGILQIVLDFTLKLFKLYFSKDISFTNILSMLEFVATYIEDQLYFMTKTKGKPTRQSDADRNERRRNAIRELQNQRDIIQKIADLFRHLQAEIIAETSAPNIISPYFRDIFSAVYHYYKHRRVPVPGHEKLTVKQYFDLIRMVIIKMKESPEYKKMQKARNGTTLTCEMYVVLFGRMFRIIITVRDGEIVLSTAYVVKRMNFNAKEGILYIKFGDGDVQK
Summary
Uniprot
ProteinModelPortal
PDB
6AR3
E-value=0.000156264,
Score=114
Ontologies
GO
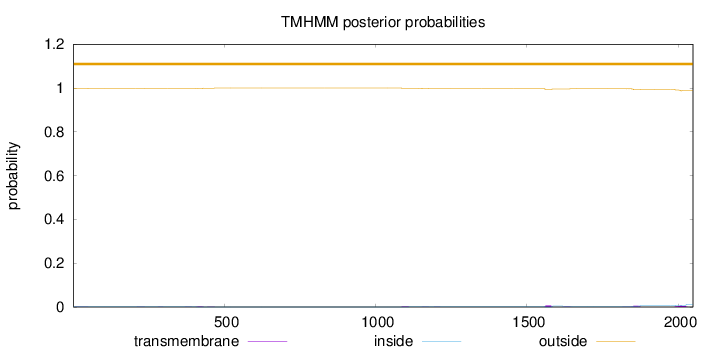
Topology
Length:
2049
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.648600000000001
Exp number, first 60 AAs:
0.01513
Total prob of N-in:
0.00316
outside
1 - 2049
Population Genetic Test Statistics
Pi
349.493608
Theta
20.890825
Tajima's D
3.265808
CLR
0.34382
CSRT
0.988800559972001
Interpretation
Uncertain
