Gene
KWMTBOMO04281
Pre Gene Modal
BGIBMGA012649
Annotation
PREDICTED:_lipase_3-like_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.088 Nuclear Reliability : 1.236
Sequence
CDS
ATGATATTTCCGAGTCTGATTTTTAAAATTGCGTTCATTTTACTCGTCGCCTTTTTAAATAAAGTTTCTAATTCACTTATAGACTCACCGATCTTGGAAAGCATTCCAGGAGACATTGCCATTGAACAAAACGATTTCGTAAAATCGTACGGTTTGGATCCGGTAGGGAATTTAGTGTCCATTGAAGTACCCGAAGACGCTAACCTTGATTTCAACGGTCTCGCCAAGAAGTACGGGCATCCGTGCGAAGAGCATTACATCGAGTCAGAGGATGGTTATATCACCAAGTTATTTCATTTACCGGGAGATAAGAAACGGCCGGTACTCATGATGCACGGCATCTTCGATTCTGCCGATACTTTTATCATCAGAGGCAACAGATCAATGGCCATATCCCTCGCGAACACTGGCTACGACGTGTGGGTGGGAAACATGAGAGGAAACAAACATTCCCGGAGACACGTTACTCTGAATCCCGACAAAGATCCTGAATTTTGGAACTTCAGTTACCACGAAATGGCGTTGTACGATCTACCTGCGACTATCGACTACATCCTGGGTCACACCGGCCAGACACGGATCTATGGTATCGGTCATTCCCTGGGCAACAATATTTTCTTTGCAATGACGTCAATGTTGCCACAATATAACAAAAAAATCAGGATTTTCATTTCGTTAGCACCGGCTGTTCACCTTGACAATATTAAATTGCCGACAAGAATACTGGCTGTGCTATCTCCCCTTAACAACTATTTGTTGAAAGGTACTGGGCAAGAGGAAATACTTGCGGATTCAACCATCCTGTTTAAAATTTTACGAAAAGTTTGTCCGATTGGTCTGCCTGGATATATTTTATGCGCTGAATTGATTATATTTTACATAGGTGGCTACGATAGGGCAGAACTAGAGCGTGAATTCTTGCCGGTCGTTATAGGTCATGTTACCAGTGGTACCTCAAGGAAAAACTTCATTCACTTGGCGCAGTCGGTGGGAGATGGGTCGGTGGATCTGTCCTCAGATGAGGAAGGCTTGCTGCCCCCCAAGAAATTACCGACCACGAGGAGAGGTCGGCAGGCTTCGAGAGGAGTGAGTGCTTCACGGCCGCGGAGGAACGCACAGGGTAGGTTCGTGTGCTCCAATACTCCGGCTGCCAGCAAAGCAGATAGTGACATGGGGGTCACTACCGAGGACCCTGGTGGCGAGAGCTGTGCCTCGCTAGGGAGCTCAAAGGCGGAGATCAACGCCGCCCGAAGAGAGCAACGCAAAGCTGTCGCGGCCGACGAAGTGTCGGAAATGGCCCGACGCGCTCGCGAGCGACGCGCTACTCTAGCGGCGGAAGGGGGGGAACCATCTGCGGTGGCCCTAAGTCAGCTTGCCTTGGACGGGGTGGACTTAGTCCTGAAGGTTGCCACCAAATCCGGCAGCCTTAAGGGCACGTTTACCCGCGGTCTGAAAGAAGCTGCCGCGGACATTAAGGAGGCGATTGGCATCCTCCTCAACAGGACGGCCTCTGACGAGGTTGCGAAGTTGCAGGAGGAAAACAGCCGTCTCCGAAATGATATGGAGGACCTCCGGCGACGAGTCACTGCGTTGAGCGAGCAGCAGCAGCGGCGTACGTCCACTGATGCCGCATCGGTAGTGGCCCCGGCTCCCGCACCGAGACCGACAAGCACTCATACCGACGACGAGGTCGAGCGCATTGTCCGGCTCTGTATGCTCCAGTGCGGGAGCATGGTCAATGCTCGTATGGAGGCAATTTCTCGGCGCCTCCCTGCGGAAATCCTCCGTCCGCCACTAGCGGCCGATACACGGCGGAGGGCTGAGGAGCCGCCGAGACCTAAGCCTAGGGAGGGGAAGCCCGCGGAGGGTGTTAAAAAGCCCGTCGAGGGAGCCCCATCGAGCGATCAGCCCACGACTGCTGGGTCTAAGGGTGAGACCTGGGTTACAGTCGTGGGCCGCAAGAAGGCACGCAGGGTCGCCAAGGCGGCCTCAGCAGCAACGCATGCCCCCGGCCAAACCGCAAAGGCGGTTGCGCAGCCTGCGCGGCGGGCTGCCAAAGGCGGTCGCAAAGGACCGGCGATACGTGCTCCGCGTTCCGAAGCTGTGACGCTCACGCTACAGCCTGGAGCTGCGGAGCGCGGCGTAACGTACCAGTCGGTCATCGCCGAAGCAAAGGCCAAGATCAAATTATCAGATCTTGGTCTTCAGGCCGTCACCCACAGGCAGGCTGCCACGGGTGCACGGTTGTTCGAGGTGGCAAGTACTACGAGTGGCAGTGCCGAAAAGGCGGACGCTCTGGCCGCCAAGATGAGGGAGGTCCTCAGCCCCGAGGACGTCCGGGTCTCCAGGCCAATGAAAACCGCAGAGGTGCGGATTACTGGCCTGGATGACTCCGTGACCTCTGAGGAGGTAGTAGCGGCCGTTGCCCGAAGTGGAGAGTGTCCATCGGACAAGGTGCGGGCCGGCGATATACGCACCGACGCCACCGGACTCGGCGTGGCCTGGGTTCGGTGCCCTGTAGCGTCGGCGAAAAAGATCGCCTTAAGCGGCAGATTGCTGAGCATCGAGATGCACTATTTCAGCTGA
Protein
MIFPSLIFKIAFILLVAFLNKVSNSLIDSPILESIPGDIAIEQNDFVKSYGLDPVGNLVSIEVPEDANLDFNGLAKKYGHPCEEHYIESEDGYITKLFHLPGDKKRPVLMMHGIFDSADTFIIRGNRSMAISLANTGYDVWVGNMRGNKHSRRHVTLNPDKDPEFWNFSYHEMALYDLPATIDYILGHTGQTRIYGIGHSLGNNIFFAMTSMLPQYNKKIRIFISLAPAVHLDNIKLPTRILAVLSPLNNYLLKGTGQEEILADSTILFKILRKVCPIGLPGYILCAELIIFYIGGYDRAELEREFLPVVIGHVTSGTSRKNFIHLAQSVGDGSVDLSSDEEGLLPPKKLPTTRRGRQASRGVSASRPRRNAQGRFVCSNTPAASKADSDMGVTTEDPGGESCASLGSSKAEINAARREQRKAVAADEVSEMARRARERRATLAAEGGEPSAVALSQLALDGVDLVLKVATKSGSLKGTFTRGLKEAAADIKEAIGILLNRTASDEVAKLQEENSRLRNDMEDLRRRVTALSEQQQRRTSTDAASVVAPAPAPRPTSTHTDDEVERIVRLCMLQCGSMVNARMEAISRRLPAEILRPPLAADTRRRAEEPPRPKPREGKPAEGVKKPVEGAPSSDQPTTAGSKGETWVTVVGRKKARRVAKAASAATHAPGQTAKAVAQPARRAAKGGRKGPAIRAPRSEAVTLTLQPGAAERGVTYQSVIAEAKAKIKLSDLGLQAVTHRQAATGARLFEVASTTSGSAEKADALAAKMREVLSPEDVRVSRPMKTAEVRITGLDDSVTSEEVVAAVARSGECPSDKVRAGDIRTDATGLGVAWVRCPVASAKKIALSGRLLSIEMHYFS
Summary
Uniprot
ProteinModelPortal
PDB
1K8Q
E-value=3.49104e-37,
Score=392
Ontologies
GO
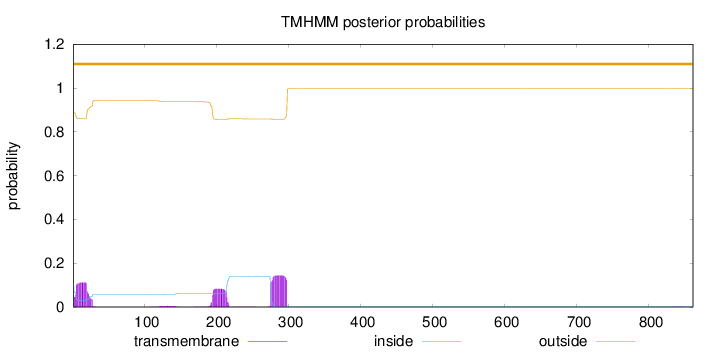
Topology
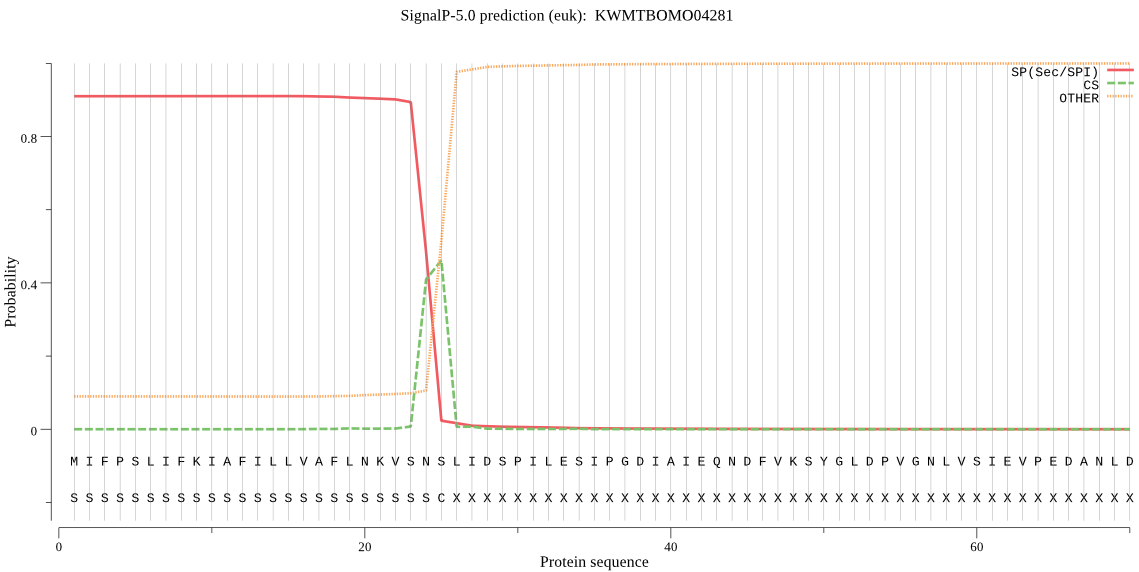
SignalP
Position: 1 - 25,
Likelihood: 0.910572
Length:
861
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.24407999999999
Exp number, first 60 AAs:
2.18553
Total prob of N-in:
0.11272
outside
1 - 861
Population Genetic Test Statistics
Pi
228.221086
Theta
168.012322
Tajima's D
0.465319
CLR
0.996472
CSRT
0.508324583770811
Interpretation
Uncertain
