Gene
KWMTBOMO04277
Annotation
PREDICTED:_phosphatidylinositol-glycan_biosynthesis_class_X_protein_isoform_X1_[Bombyx_mori]
Full name
Phosphatidylinositol-glycan biosynthesis class X protein
Location in the cell
PlasmaMembrane Reliability : 2.275
Sequence
CDS
ATGTTGACAAATATTTTTAGTTTTCGAATATGTATCGTTGTTTTTGTTGCAATCAAAGTTTTTTCGGCCAGTAAATGTGATTATGATGTGAAAATAGCTCAGATACTCATCAATGATGGATATCACAGGCATCATTACAGGAACCTAACCTACCACATAATTTTCAATGAGGATTCAGAGTCAAAAGGTATATACAATGGATGCAGAATTGGCCTCGACCAGTACCTGCCTGCTGGGGTGTTTGTTAATCCTAATGAACTGAATGAGTTCAATCGTCTGGAAAGGTTGAATGCATTTCCGAGGGGTCGGGTCAACATAGAGCTACCAACTGAAGAAGCAGAACCTATAAATGTGTTCATTATAAGTGAATTGACTGATGATGAAATACAAATGTGGCTACCAGTTCATGCTCGGTATCACGAATCTGTATCTGGTGGAGGGATGGTACAGTATGACATAGGACCCCCGAAAATGTACCTATATTGTGCTGATCGTAGACTGGACGTGTGCGACCGCAGTGTGTCACCATCTGTCACTTTGACATGTAACGGTGGTGTCAAGATGTGCTCGTGGAGAGATATACCTTACACTGTGTTGACAGATCCTCTCGTGTGGCGCGTTCCAGTCGGGAACTCCAACCACTTCTACATCATCGGCATAGGTACAGCATTCGCCATAATGGTCGGCACCACAATGGTGATCAGAGCGATGCATGACCACATTTTGAAGCACCGACGTCTGATTATACGATGA
Protein
MLTNIFSFRICIVVFVAIKVFSASKCDYDVKIAQILINDGYHRHHYRNLTYHIIFNEDSESKGIYNGCRIGLDQYLPAGVFVNPNELNEFNRLERLNAFPRGRVNIELPTEEAEPINVFIISELTDDEIQMWLPVHARYHESVSGGGMVQYDIGPPKMYLYCADRRLDVCDRSVSPSVTLTCNGGVKMCSWRDIPYTVLTDPLVWRVPVGNSNHFYIIGIGTAFAIMVGTTMVIRAMHDHILKHRRLIIR
Summary
Description
Essential component of glycosylphosphatidylinositol-mannosyltransferase 1 which transfers the first of the 4 mannoses in the GPI-anchor precursors during GPI-anchor biosynthesis. Probably acts by stabilizing the mannosyltransferase PIGM (By similarity).
Subunit
Interacts with PIGM.
Similarity
Belongs to the PIGX family.
Keywords
Alternative splicing
Complete proteome
Endoplasmic reticulum
Glycoprotein
GPI-anchor biosynthesis
Membrane
Reference proteome
Signal
Transmembrane
Transmembrane helix
Feature
chain Phosphatidylinositol-glycan biosynthesis class X protein
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A2W1BMQ1
A0A2H1VC44
A0A194RGK0
A0A194Q192
A0A1E1W9Q6
A0A1B6BZ57
+ More
A0A1B6EKN5 A0A1B6KZA0 A0A1Y1K8M4 A0A0V0GBL8 A0A3Q0JKE3 A0A1S4E713 A0A069DQK5 T1I1R7 A0A067QQI7 A0A2P8YBV0 E2AE44 A0A088AJP6 A0A2G9QKP5 H3A3C4 A0A0L0CP74 A0A154PBW0 F4WEN9 B4KQ38 K9IR82 A0A026W3C6 A0A0K8SN19 Q28YB4 B4GD50 A0A0A9YDD3 A0A195FM55 J3JWE6 A0A195E7W8 A0A2R7WCY1 A0A232F0S5 A0A195C664 A0A3B0J6T6 A0A146M4Y8 G1Q8N8 W5JW65 K7IRT5 A0A0L7R1F2 F6RKI9 U6DYQ3 M3XXL1 A0A0M4EEZ5 A0A2Y9KNF1 N6STZ1 A0A2Y9SMT5 S7N4P1 A0A2M4CMR8 B4P255 A0A2M4BY56 A0A084VL22 A0A2Y9NY62 G3T1M3 G3UEV2 A0A3Q7NM60 G1LAP0 A0A182MVH5 A0A2K5ZZL4 A0A341C037 A0A091KA67 A0A1A9XIV1 A0A384DHW4 B4MR01 A0A383Z9I2 M3WNE3 A0A1W4WB87 A0A1I8M2T3 G5ALC7 A0A195AVL9 A0A158NF05 B3N9C6 A0A099ZS15 A0A1U8DWA6 A0A3Q0FT40 A0A3Q7V1X3 A0A091DLV8 A0A3Q0FPT1 A0A2Y9HF86 A0A087WSN5 F6YX19 A0A182JK25 A0A2U3YGU5 A0A2U4A9P9 F7DFD7 Q99LV7 A0A1Q3F3K8 A0A2K6GSB4 H9ZAK0 F1LRM6 A0A1J1J015 A0A2J8RG78 H9F2G9 A0A182MZ47
A0A1B6EKN5 A0A1B6KZA0 A0A1Y1K8M4 A0A0V0GBL8 A0A3Q0JKE3 A0A1S4E713 A0A069DQK5 T1I1R7 A0A067QQI7 A0A2P8YBV0 E2AE44 A0A088AJP6 A0A2G9QKP5 H3A3C4 A0A0L0CP74 A0A154PBW0 F4WEN9 B4KQ38 K9IR82 A0A026W3C6 A0A0K8SN19 Q28YB4 B4GD50 A0A0A9YDD3 A0A195FM55 J3JWE6 A0A195E7W8 A0A2R7WCY1 A0A232F0S5 A0A195C664 A0A3B0J6T6 A0A146M4Y8 G1Q8N8 W5JW65 K7IRT5 A0A0L7R1F2 F6RKI9 U6DYQ3 M3XXL1 A0A0M4EEZ5 A0A2Y9KNF1 N6STZ1 A0A2Y9SMT5 S7N4P1 A0A2M4CMR8 B4P255 A0A2M4BY56 A0A084VL22 A0A2Y9NY62 G3T1M3 G3UEV2 A0A3Q7NM60 G1LAP0 A0A182MVH5 A0A2K5ZZL4 A0A341C037 A0A091KA67 A0A1A9XIV1 A0A384DHW4 B4MR01 A0A383Z9I2 M3WNE3 A0A1W4WB87 A0A1I8M2T3 G5ALC7 A0A195AVL9 A0A158NF05 B3N9C6 A0A099ZS15 A0A1U8DWA6 A0A3Q0FT40 A0A3Q7V1X3 A0A091DLV8 A0A3Q0FPT1 A0A2Y9HF86 A0A087WSN5 F6YX19 A0A182JK25 A0A2U3YGU5 A0A2U4A9P9 F7DFD7 Q99LV7 A0A1Q3F3K8 A0A2K6GSB4 H9ZAK0 F1LRM6 A0A1J1J015 A0A2J8RG78 H9F2G9 A0A182MZ47
Pubmed
28756777
26354079
28004739
26334808
24845553
29403074
+ More
20798317 9215903 26108605 21719571 17994087 24508170 30249741 15632085 25401762 22516182 28648823 26823975 21993624 20920257 23761445 20075255 19892987 23537049 17550304 24438588 20010809 17975172 25315136 21993625 21347285 19468303 16141072 15489334 15635094 25319552 15057822
20798317 9215903 26108605 21719571 17994087 24508170 30249741 15632085 25401762 22516182 28648823 26823975 21993624 20920257 23761445 20075255 19892987 23537049 17550304 24438588 20010809 17975172 25315136 21993625 21347285 19468303 16141072 15489334 15635094 25319552 15057822
EMBL
KZ150021
PZC74894.1
ODYU01001758
SOQ38397.1
KQ460211
KPJ16569.1
+ More
KQ459580 KPI99352.1 GDQN01007375 JAT83679.1 GEDC01030747 GEDC01024603 GEDC01002031 JAS06551.1 JAS12695.1 JAS35267.1 GECZ01031280 GECZ01018174 JAS38489.1 JAS51595.1 GEBQ01023200 JAT16777.1 GEZM01096141 JAV55127.1 GECL01000636 JAP05488.1 GBGD01002923 JAC85966.1 ACPB03018229 ACPB03018230 KK853091 KDR11544.1 PYGN01000719 PSN41742.1 GL438827 EFN68277.1 KV961180 PIO16164.1 AFYH01210524 AFYH01210525 AFYH01210526 JRES01000105 KNC34071.1 KQ434858 KZC08904.1 GL888103 EGI67402.1 CH933808 EDW08140.1 GABZ01002455 JAA51070.1 KK107455 QOIP01000010 EZA50528.1 RLU17700.1 GBRD01011082 GBRD01011081 JAG54742.1 CM000071 EAL26051.3 CH479181 EDW32545.1 GBHO01013980 GBHO01013979 GBHO01013978 JAG29624.1 JAG29625.1 JAG29626.1 KQ981490 KYN41337.1 BT127564 AEE62526.1 KQ979479 KYN21310.1 KK854561 PTY16890.1 NNAY01001375 OXU24192.1 KQ978287 KYM95666.1 OUUW01000001 SPP75552.1 GDHC01004140 JAQ14489.1 AAPE02025167 ADMH02000203 ETN67380.1 KQ414668 KOC64621.1 HAAF01015367 CCP87187.1 AEYP01017513 AEYP01017514 AEYP01017515 CP012524 ALC42740.1 APGK01056839 KB741277 KB632028 ENN71154.1 ERL88115.1 KE163428 EPQ12029.1 GGFL01002387 MBW66565.1 CM000157 EDW89256.2 GGFJ01008801 MBW57942.1 ATLV01014397 KE524970 KFB38666.1 ACTA01042684 ACTA01050684 ACTA01058684 AXCM01007649 KK502098 KFP20651.1 CH963849 EDW74540.1 AANG04001815 JH165776 EHA97837.1 KQ976736 KYM76070.1 ADTU01013693 CH954177 EDV59613.2 KL896584 KGL83555.1 KN122104 KFO33119.1 AC135962 AK008583 BC002202 GFDL01012893 JAV22152.1 JU476162 AFH32966.1 AABR07034444 CVRI01000061 CRL04185.1 NDHI03003699 PNJ07506.1 PNJ07509.1 JU325072 AFE68828.1
KQ459580 KPI99352.1 GDQN01007375 JAT83679.1 GEDC01030747 GEDC01024603 GEDC01002031 JAS06551.1 JAS12695.1 JAS35267.1 GECZ01031280 GECZ01018174 JAS38489.1 JAS51595.1 GEBQ01023200 JAT16777.1 GEZM01096141 JAV55127.1 GECL01000636 JAP05488.1 GBGD01002923 JAC85966.1 ACPB03018229 ACPB03018230 KK853091 KDR11544.1 PYGN01000719 PSN41742.1 GL438827 EFN68277.1 KV961180 PIO16164.1 AFYH01210524 AFYH01210525 AFYH01210526 JRES01000105 KNC34071.1 KQ434858 KZC08904.1 GL888103 EGI67402.1 CH933808 EDW08140.1 GABZ01002455 JAA51070.1 KK107455 QOIP01000010 EZA50528.1 RLU17700.1 GBRD01011082 GBRD01011081 JAG54742.1 CM000071 EAL26051.3 CH479181 EDW32545.1 GBHO01013980 GBHO01013979 GBHO01013978 JAG29624.1 JAG29625.1 JAG29626.1 KQ981490 KYN41337.1 BT127564 AEE62526.1 KQ979479 KYN21310.1 KK854561 PTY16890.1 NNAY01001375 OXU24192.1 KQ978287 KYM95666.1 OUUW01000001 SPP75552.1 GDHC01004140 JAQ14489.1 AAPE02025167 ADMH02000203 ETN67380.1 KQ414668 KOC64621.1 HAAF01015367 CCP87187.1 AEYP01017513 AEYP01017514 AEYP01017515 CP012524 ALC42740.1 APGK01056839 KB741277 KB632028 ENN71154.1 ERL88115.1 KE163428 EPQ12029.1 GGFL01002387 MBW66565.1 CM000157 EDW89256.2 GGFJ01008801 MBW57942.1 ATLV01014397 KE524970 KFB38666.1 ACTA01042684 ACTA01050684 ACTA01058684 AXCM01007649 KK502098 KFP20651.1 CH963849 EDW74540.1 AANG04001815 JH165776 EHA97837.1 KQ976736 KYM76070.1 ADTU01013693 CH954177 EDV59613.2 KL896584 KGL83555.1 KN122104 KFO33119.1 AC135962 AK008583 BC002202 GFDL01012893 JAV22152.1 JU476162 AFH32966.1 AABR07034444 CVRI01000061 CRL04185.1 NDHI03003699 PNJ07506.1 PNJ07509.1 JU325072 AFE68828.1
Proteomes
UP000053240
UP000053268
UP000079169
UP000015103
UP000027135
UP000245037
+ More
UP000000311 UP000005203 UP000008672 UP000037069 UP000076502 UP000007755 UP000009192 UP000053097 UP000279307 UP000001819 UP000008744 UP000078541 UP000078492 UP000215335 UP000078542 UP000268350 UP000001074 UP000000673 UP000002358 UP000053825 UP000002281 UP000000715 UP000092553 UP000248482 UP000019118 UP000030742 UP000002282 UP000030765 UP000248483 UP000007646 UP000286641 UP000008912 UP000075883 UP000233140 UP000252040 UP000053119 UP000092443 UP000261680 UP000007798 UP000261681 UP000011712 UP000192221 UP000095301 UP000006813 UP000078540 UP000005205 UP000008711 UP000053641 UP000189705 UP000286642 UP000028990 UP000248481 UP000000589 UP000075880 UP000245341 UP000245320 UP000233160 UP000002494 UP000183832 UP000075884
UP000000311 UP000005203 UP000008672 UP000037069 UP000076502 UP000007755 UP000009192 UP000053097 UP000279307 UP000001819 UP000008744 UP000078541 UP000078492 UP000215335 UP000078542 UP000268350 UP000001074 UP000000673 UP000002358 UP000053825 UP000002281 UP000000715 UP000092553 UP000248482 UP000019118 UP000030742 UP000002282 UP000030765 UP000248483 UP000007646 UP000286641 UP000008912 UP000075883 UP000233140 UP000252040 UP000053119 UP000092443 UP000261680 UP000007798 UP000261681 UP000011712 UP000192221 UP000095301 UP000006813 UP000078540 UP000005205 UP000008711 UP000053641 UP000189705 UP000286642 UP000028990 UP000248481 UP000000589 UP000075880 UP000245341 UP000245320 UP000233160 UP000002494 UP000183832 UP000075884
Pfam
Interpro
IPR040039
PIGX
+ More
IPR013233 PIG-X/PBN1
IPR027417 P-loop_NTPase
IPR030547 XRCC2
IPR020588 RecA_ATP-bd
IPR013632 DNA_recomb/repair_Rad51_C
IPR012676 TGS-like
IPR033728 ThrRS_core
IPR004154 Anticodon-bd
IPR006195 aa-tRNA-synth_II
IPR018163 Thr/Ala-tRNA-synth_IIc_edit
IPR004095 TGS
IPR002320 Thr-tRNA-ligase_IIa
IPR002314 aa-tRNA-synt_IIb
IPR012675 Beta-grasp_dom_sf
IPR036621 Anticodon-bd_dom_sf
IPR012947 tRNA_SAD
IPR013233 PIG-X/PBN1
IPR027417 P-loop_NTPase
IPR030547 XRCC2
IPR020588 RecA_ATP-bd
IPR013632 DNA_recomb/repair_Rad51_C
IPR012676 TGS-like
IPR033728 ThrRS_core
IPR004154 Anticodon-bd
IPR006195 aa-tRNA-synth_II
IPR018163 Thr/Ala-tRNA-synth_IIc_edit
IPR004095 TGS
IPR002320 Thr-tRNA-ligase_IIa
IPR002314 aa-tRNA-synt_IIb
IPR012675 Beta-grasp_dom_sf
IPR036621 Anticodon-bd_dom_sf
IPR012947 tRNA_SAD
Gene 3D
ProteinModelPortal
A0A2W1BMQ1
A0A2H1VC44
A0A194RGK0
A0A194Q192
A0A1E1W9Q6
A0A1B6BZ57
+ More
A0A1B6EKN5 A0A1B6KZA0 A0A1Y1K8M4 A0A0V0GBL8 A0A3Q0JKE3 A0A1S4E713 A0A069DQK5 T1I1R7 A0A067QQI7 A0A2P8YBV0 E2AE44 A0A088AJP6 A0A2G9QKP5 H3A3C4 A0A0L0CP74 A0A154PBW0 F4WEN9 B4KQ38 K9IR82 A0A026W3C6 A0A0K8SN19 Q28YB4 B4GD50 A0A0A9YDD3 A0A195FM55 J3JWE6 A0A195E7W8 A0A2R7WCY1 A0A232F0S5 A0A195C664 A0A3B0J6T6 A0A146M4Y8 G1Q8N8 W5JW65 K7IRT5 A0A0L7R1F2 F6RKI9 U6DYQ3 M3XXL1 A0A0M4EEZ5 A0A2Y9KNF1 N6STZ1 A0A2Y9SMT5 S7N4P1 A0A2M4CMR8 B4P255 A0A2M4BY56 A0A084VL22 A0A2Y9NY62 G3T1M3 G3UEV2 A0A3Q7NM60 G1LAP0 A0A182MVH5 A0A2K5ZZL4 A0A341C037 A0A091KA67 A0A1A9XIV1 A0A384DHW4 B4MR01 A0A383Z9I2 M3WNE3 A0A1W4WB87 A0A1I8M2T3 G5ALC7 A0A195AVL9 A0A158NF05 B3N9C6 A0A099ZS15 A0A1U8DWA6 A0A3Q0FT40 A0A3Q7V1X3 A0A091DLV8 A0A3Q0FPT1 A0A2Y9HF86 A0A087WSN5 F6YX19 A0A182JK25 A0A2U3YGU5 A0A2U4A9P9 F7DFD7 Q99LV7 A0A1Q3F3K8 A0A2K6GSB4 H9ZAK0 F1LRM6 A0A1J1J015 A0A2J8RG78 H9F2G9 A0A182MZ47
A0A1B6EKN5 A0A1B6KZA0 A0A1Y1K8M4 A0A0V0GBL8 A0A3Q0JKE3 A0A1S4E713 A0A069DQK5 T1I1R7 A0A067QQI7 A0A2P8YBV0 E2AE44 A0A088AJP6 A0A2G9QKP5 H3A3C4 A0A0L0CP74 A0A154PBW0 F4WEN9 B4KQ38 K9IR82 A0A026W3C6 A0A0K8SN19 Q28YB4 B4GD50 A0A0A9YDD3 A0A195FM55 J3JWE6 A0A195E7W8 A0A2R7WCY1 A0A232F0S5 A0A195C664 A0A3B0J6T6 A0A146M4Y8 G1Q8N8 W5JW65 K7IRT5 A0A0L7R1F2 F6RKI9 U6DYQ3 M3XXL1 A0A0M4EEZ5 A0A2Y9KNF1 N6STZ1 A0A2Y9SMT5 S7N4P1 A0A2M4CMR8 B4P255 A0A2M4BY56 A0A084VL22 A0A2Y9NY62 G3T1M3 G3UEV2 A0A3Q7NM60 G1LAP0 A0A182MVH5 A0A2K5ZZL4 A0A341C037 A0A091KA67 A0A1A9XIV1 A0A384DHW4 B4MR01 A0A383Z9I2 M3WNE3 A0A1W4WB87 A0A1I8M2T3 G5ALC7 A0A195AVL9 A0A158NF05 B3N9C6 A0A099ZS15 A0A1U8DWA6 A0A3Q0FT40 A0A3Q7V1X3 A0A091DLV8 A0A3Q0FPT1 A0A2Y9HF86 A0A087WSN5 F6YX19 A0A182JK25 A0A2U3YGU5 A0A2U4A9P9 F7DFD7 Q99LV7 A0A1Q3F3K8 A0A2K6GSB4 H9ZAK0 F1LRM6 A0A1J1J015 A0A2J8RG78 H9F2G9 A0A182MZ47
Ontologies
PATHWAY
GO
Topology
Subcellular location
Endoplasmic reticulum membrane
SignalP
Position: 1 - 22,
Likelihood: 0.914401
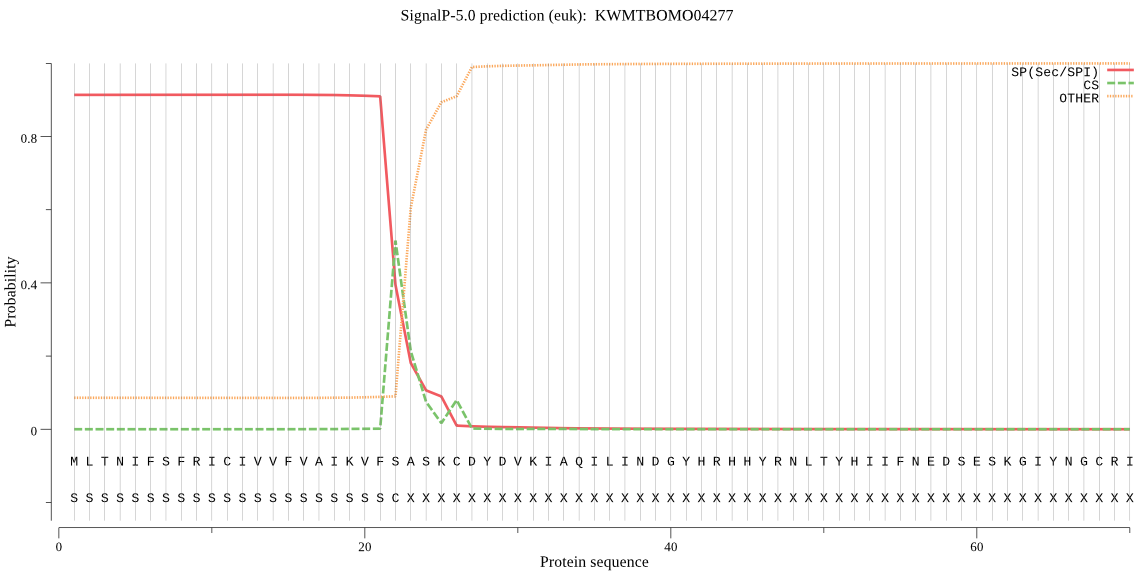
Length:
250
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
36.36377
Exp number, first 60 AAs:
14.0409
Total prob of N-in:
0.14681
POSSIBLE N-term signal
sequence
outside
1 - 214
TMhelix
215 - 237
inside
238 - 250
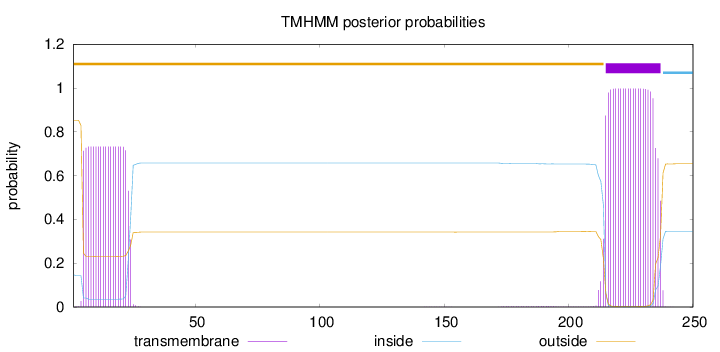
Population Genetic Test Statistics
Pi
217.377977
Theta
171.996048
Tajima's D
0.920989
CLR
0
CSRT
0.637618119094045
Interpretation
Uncertain
