Gene
KWMTBOMO04271 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014227
Annotation
PREDICTED:_myosin_heavy_chain?_muscle_isoform_X8_[Papilio_xuthus]
Location in the cell
Cytoskeletal Reliability : 4.443
Sequence
CDS
ATGCCGAAACCTGTCGTCCAAGAGGGCGAGGACCCCGATCCGACCCCGTACCTGTTCGTTTCTCTGGAACAGAAGCGTATCGACCAGAGCAAACCTTACGATGGCAAGAAGGCATGCTGGGTGCCTGATGAAAAGGAGGGCTTCGTGCAGGGCGAGATCAAGGCCACCAAGGGAGACCTGGTCACCGTCAACCTGCCCGGTGGCGAGACTAAAGACTTCAAAAAGGATCAAGTAGCGCAAGTCAACCCGCCCAAGTACGAAAAATGCGAAGATATGTCCAACTTGACATATCTCAACGACGCTTCAGTATTGTATAATTTGAAGCAGAGATATTACCATAAACTCATTTACACGTACTCGGGTCTCTTCTGTGTGGCCATCAACCCTTACAAGAGATTCCCCGTGTACACGACACGATGCGCCAAGCTGTACCGTGGCAAGCGTCGTTCGGAAGTGCCACCTCACATCTTCGCCATTTCCGACGGCGCTTACGTCAACATGCTGACCAACCACGAGAATCAATCTATGTTGATTACCGGTGAGTCTGGTGCTGGTAAGACTGAGAACACGAAGAAGGTAATTGCGTACTTCGCCACTGTCGGTGCCTCCCAGAAGAAGGACCCAAGCCAGGAGAAGAAGGGCTCCCTGGAAGATCAAGTAGTACAAACCAACCCTGTACTTGAAGCGTTCGGTAACGCTAAGACCGTCCGTAACGACAACTCCTCTCGTTTCGGTAAATTCATCCGTATCCATTTCGGTCCATCCGGAAAACTGGCTGGTGCTGACATCGAAACCTATCTGCTTGAGAAGGCTCGTGTCATCTCCCAGCAAGCTCTTGAGCGTTCTTACCACATCTTCTACCAGATGATGTCCGGCTCCGTCAACGGTCTTAAAGCCATCTGCCTTTTGTCCAACGACGTCATGGACTACAACATCGTGTCCCAAGGAAAGACGGTCATTCCCGGCGTCGATGACGGGGAGGAAATGAGAATTACCGACCAAGCCTTCGACATCCTTGGTTTCACCCAAGAGGAGAAAGACAATGTATACAAGATCACTGCTGCCGTCATGCATATGGGCTGCATGAAGTTCAAGCAGAGGGGTCGCGAGGAGCAGGCCGAGGCTGACGGTACCGAGGACGGTGACAAAGTTGCCAAGCTCCTCGGCGTCGACTGCGCTGACCTGTACAAGAACTTGTTGAAGCCCCGCATCAAGGTCGGAAACGAGTTCGTGACCCAGGGTCGTAACAAGGACCAGGTCATCAACTCCGTTGGTGCCCTCTGCAAGGGTATCTTCGATCGTCTCTTCAAGTGGCTCGTCAAGAAATGTAACGAGACCCTAGACACCAAACAAAAGAGGCAGCACTTCATCGGTGTACTGGATATTGCTGGTTTCGAAATTTTCGACTTCAACGGTTTTGAGCAACTCTGCATTAACTTCACAAACGAGAAACTGCAGCAGTTCTTTAACCATCACATGTTCGTGCTCGAGCAAGAGGAGTACCAGCGCGAAGGCATCGAATGGACTTTCATTGATTTTGGCATGGACCTCCAAAATTGCATTGACCTTATTGAAAAGCCTATGGGTATCCTCTCCATTCTTGAGGAAGAGTCTATGTTCCCGAAAGCCACCGACCAGACCTTCGTTGAGAAGCTGAACAACAACCACTTGGGCAAATCTGCTCCTTACTTGAAGCCCAAACCCCCCAAGCCAGGTTGCCAAGCTGCTCACTTCGCCATTGGTCACTACGCCGGTAACGTCGGCTACAACATCACCGGTTGGCTCGAGAAGAACAAGGACCCGCTTAACGACACCGTCGTCGACCAGTTCAAGAAGGGCCAGAACAAACTTCTGGTCGAGATCTTCGCTGATCATCCTGGCCAGTCTGGTGATGCTTCCGGCGGTGCCGGTGGCAAGGGCGCTGGTGGCAAACGCGCTAAGGGTTCTGCCTTCCAGACTGTATCATCACTCTACAGGGAACAACTTAACAACTTGATGACGACGCTGAGATCCACCCAACCTCACTTCGTGCGTTGTATCATTCCCAATGAATTGAAACAGGCTGGTCTCATCGACTCTCACCTTGTGATGCATCAGCTCACCTGTAACGGTGTGCTTGAAGGCATCCGTATTTGCCGTAAAGGTTTCCCCAACAGAATGGTATACCCTGACTTCAAGCTCCGATACAAAATTCTGTGCCCGAACCTCATCAAAGAACCTATCACACCGGAGAAAGCCACCGAGAAAATTCTCGAACAAACTGGTCTGGATTCGGAATCTTTCAGGCTCGGAAAGACAAAGGTATTCTTCCGCGCTGGTGTCCTGGGTCAGATGGAAGAGCTGCGTGATGACAGGCTCTCCAAGATCGTATCATGGCTCCAGGCTTACATCCGCGGTTACTTGTCCCGTAAGGAGTACAAGAAGCTGCAGGAGCAGAGATTGGCTCTCCAAGTTGTCCAGCGCAACTTGCGCAAGTACCTGCAACTCCGCACCTGGCCCTGGTGGAAACTGTGGCAGAAGGTCAAGCCTCTCCTCAACGTCACCCGCATCGAGGACGAGATCGCGAAACTCGAGGAGAAGGCACAGAAGGCTCAGGAGGCTTTCGAGAAGGAGGAGAAGCTCCGCAAGGAGGTCGAGGCCCTCAACGCCAAGCTGCTTGAGGAGAAGACCGCCCTGCTCGCCAACCTCGAAGGAAACCAGGGCAGCCTTGCCGAGACCCAGGAGCGCGCTAACAAGCTCCAGGCCCAGAAGGCTGATCTCGAGAACCAACTCAGGGACACCCAAGACCGCCTCACCCAGGAAGAGGATGCCCGTAACCAGCTCTTCCAAGCTAAGAAGAAGCTCGAACAGGAAGTCTCTGGCCTCAAGAAGGATGTCGAGGACTTGGAACTGTCCGTACAGAAGTCCGAACAGGACAAGGCTACCAAGGACCACCAGATCCGCAACTTGAATGATGAGATCGCTCACCAGGATGAGCTCATCAACAAGTTGAACAAGGAGAAGAAACTCCAAGGCGAGACCAACCAGAAGACCTCAGAAGAACTCCAGGCTGCCGAAGACAAGGTCAACCACCTTAACAAGGTCAAGCAAAAGTTGGAGCAGACCCTTGACGAGCTCGAAGACTCTCTGGAGCGCGAAAAGAAACTGCGTGCTGACGTCGAAAAGCAGAGGAGGAAGGTTGAGGGAGACCTCAAGCTCACCCAGGAAGCTGTCGCCGACCTCGAGCGCAACAAGAAGGAACTCGAACAGACCATCCAACGCAAGGACAAGGAAATCTCATCTCTCACCGCCAAACTCGAAGACGAACAGTCCCTGGTCAGCAAGCTCCAGAAACAGATCAAGGAGCTCCAAGGCCGCATCGAGGAGCTCGAAGAGGAAGTCGAGAGCGAGCGCCAGGCTCGCGCTAAGGCTGAGAAACAACGCGCTGACCTCGCTCGTGAACTTGAAGAGCTCGGCGAAAGGCTCGAGGAAGCCGGTGGCGCCACCTCTGCTCAGATCGAACTCAACAAGAAGCGCGAGGCCGAGCTTAGCAAGCTGCGCCGTGACCTCGAGGAGGCCAACATCCAGCACGAAGCCACCCTCGCCAACCTCCGCAAGAAGCACAACGATGCCGTCTCCGAGATGGGTGAGCAGCTCGACCAGCTCAACAAGCTCAAGGCTAAGGCTGAGAAAGAGCGTGCTCAATACTTTAGCGAAGTCAATGACCTCCGTGCCGGTCTCGACCACTTGTCCAACGAAAAGGCTGCACAAGAAAAGATCGTCAAGCAACTGCAGCACCAGCTCAACGAAGTCCAGAGCAAGGCTGACGAAGCCAACCGCACCCTCAACGACCTGGATGCCGCTAAGAAGAAGCTGTCCATCGAGAACTCCGACCTTCTCCGCCAACTGGAGGAGGCCGAGTCCCAGGTGTCTCAGCTCTCCAAGATCAAGGTGTCGCTCACCACACAGCTCGAAGACACCAAGAGGCTCGCCGACGAGGAAGCCAGGGAACGTGCTACCCTTCTTGGCAAGTTCCGCAACTTGGAACACGACTTGGACAACATCCGCGAACAGGTCGAGGAGGAAGCCGAGGGCAAGGCTGATCTTCAACGCCAACTGTCCAAGGCCAACGCTGAGGCTCAACTGTGGCGCTCCAAGTACGAGTCCGAGGGCGTCGCTCGCTCCGAGGAACTCGAAGAGGCCAAGCGCAAGCTCCAGGCTCGTCTCGCCGAAGCCGAGGAGACAATCGAATCCCTCAACCAGAAGGTTGTCGCTCTCGAGAAGACCAAGCAGCGTCTCGCCACCGAGGTCGAGGACCTGCAGCTGGAGGTCGACCGTGCCACTGCTATTGCCAACGCTGCAGAAAAGAAACAGAAGGCCTTCGACAAAATCATCGGAGAATGGAAACTCAAGGTCGACGACCTTGCTGCCGAACTCGATGCCAGCCAGAAGGAATGCCGCAACTACTCCACCGAATTGTTCCGCCTCAAGGGTGCCTACGAAGAAGGCCAGGAACAGCTCGAGGCTGTCCGCCGCGAGAACAAGAACCTTGCCGACGAAGTCAAGGACCTCCTGGACCAGATCGGCGAAGGTGGCCGCAACATCCACGAAATCGAGAAAGCCAGGAAGCGTCTCGAGGCCGAGAAGGACGAGCTCCAGGCCGCCCTCGAAGAGGCCGAAGCGGCTCTCGAACAAGAAGAGAACAAGGTCCTCCGCGCTCAGCTCGAGCTGTCTCAAGTCAGACAGGAGATCGACAGACGCATCCAAGAGAAGGAGGAAGAATTCGAAAACACACGCAAGAACCACCAGCGTGCCCTCGACTCCATGCAGGCTTCCCTCGAAGCCGAGGCTAAGGGCAAGGCTGAGGCGTTGCGCATGAAGAAGAAGCTCGAGGCAGACATCAACGAGCTTGAAATCGCCCTCGACCACGCCAACAAGGCTAACGCCGAGGCCCAGAAGAACATCAAACGCTACCAGGCACAAATCAAGGATCTGCAAACCGCTCTCGAGGAGGAGCAGCGCGCCCGTGACGATGCCCGCGAACAGCTCGGCATCTCGGAGCGTCGCGCCAATGCTCTTCAGAACGAACTGGAAGAGTCCCGCACACTCTTGGAGCAGGCCGACCGCGCCCGCCGCCAGGCCGAGCAGGAACTCAGTGACGCCCACGAGCAACTCAACGAGCTCTCCGCACAGAGCGCTTCCCTCTCTGCCGCCAAGAGGAAACTCGAGTCCGAGCTGCAAACCCTGCACTCCGACCTCGACGAACTTCTCAACGAGGCCAAGAACTCCGAAGAGAAGGCTAAGAAGGCCATGGTCGACGCCGCTCGTCTGGCGGACGAGCTCCGCGCCGAGCAGGATCACGCTCAGACCCAGGAGAAACTCCGCAAGGCTCTCGAGCAACAAATCAAGGAACTCCAAGTTAGGCTCGATGAGGCCGAGGCCAACGCGCTCAAGGGAGGCAAGAAGGCCATCCAGAAACTCGAACAGAGAGTCAGAGAACTCGAGAACGAGCTCGACGGCGAACAGAGGAGGCACGCTGACGCACAGAAGAACTTGCGCAAGTCCGAGAGGCGCATCAAGGAGCTCACCTTCCAGGCAGAAGAAGACCGCAAGAACCACGAACGTATGCAGGACCTCGTCGATAAACTGCAGCAGAAGATCAAGACATACAAGAGGCAGATCGAGGAGGCCGAAGAGATTGCCGCCCTCAACTTGGCTAAGTTCCGCAAGGCGCAACAGGAGCTCGAGGAGGCCGAAGAGAGGGCAGACCTCGCCGAGCAGGCCATCAGCAAGTTCCGCGGCAAGGGACGCGCGGGATCTGCAGCGAGAGGAGTCAGCCCGGCGCCCCAGCGCTCGCGCCCCGCCCTTGCTGACGGCTTCGGCACCTTCCCACCTAGGTTCGACCTGGCGCCCGAAGATTTCTAA
Protein
MPKPVVQEGEDPDPTPYLFVSLEQKRIDQSKPYDGKKACWVPDEKEGFVQGEIKATKGDLVTVNLPGGETKDFKKDQVAQVNPPKYEKCEDMSNLTYLNDASVLYNLKQRYYHKLIYTYSGLFCVAINPYKRFPVYTTRCAKLYRGKRRSEVPPHIFAISDGAYVNMLTNHENQSMLITGESGAGKTENTKKVIAYFATVGASQKKDPSQEKKGSLEDQVVQTNPVLEAFGNAKTVRNDNSSRFGKFIRIHFGPSGKLAGADIETYLLEKARVISQQALERSYHIFYQMMSGSVNGLKAICLLSNDVMDYNIVSQGKTVIPGVDDGEEMRITDQAFDILGFTQEEKDNVYKITAAVMHMGCMKFKQRGREEQAEADGTEDGDKVAKLLGVDCADLYKNLLKPRIKVGNEFVTQGRNKDQVINSVGALCKGIFDRLFKWLVKKCNETLDTKQKRQHFIGVLDIAGFEIFDFNGFEQLCINFTNEKLQQFFNHHMFVLEQEEYQREGIEWTFIDFGMDLQNCIDLIEKPMGILSILEEESMFPKATDQTFVEKLNNNHLGKSAPYLKPKPPKPGCQAAHFAIGHYAGNVGYNITGWLEKNKDPLNDTVVDQFKKGQNKLLVEIFADHPGQSGDASGGAGGKGAGGKRAKGSAFQTVSSLYREQLNNLMTTLRSTQPHFVRCIIPNELKQAGLIDSHLVMHQLTCNGVLEGIRICRKGFPNRMVYPDFKLRYKILCPNLIKEPITPEKATEKILEQTGLDSESFRLGKTKVFFRAGVLGQMEELRDDRLSKIVSWLQAYIRGYLSRKEYKKLQEQRLALQVVQRNLRKYLQLRTWPWWKLWQKVKPLLNVTRIEDEIAKLEEKAQKAQEAFEKEEKLRKEVEALNAKLLEEKTALLANLEGNQGSLAETQERANKLQAQKADLENQLRDTQDRLTQEEDARNQLFQAKKKLEQEVSGLKKDVEDLELSVQKSEQDKATKDHQIRNLNDEIAHQDELINKLNKEKKLQGETNQKTSEELQAAEDKVNHLNKVKQKLEQTLDELEDSLEREKKLRADVEKQRRKVEGDLKLTQEAVADLERNKKELEQTIQRKDKEISSLTAKLEDEQSLVSKLQKQIKELQGRIEELEEEVESERQARAKAEKQRADLARELEELGERLEEAGGATSAQIELNKKREAELSKLRRDLEEANIQHEATLANLRKKHNDAVSEMGEQLDQLNKLKAKAEKERAQYFSEVNDLRAGLDHLSNEKAAQEKIVKQLQHQLNEVQSKADEANRTLNDLDAAKKKLSIENSDLLRQLEEAESQVSQLSKIKVSLTTQLEDTKRLADEEARERATLLGKFRNLEHDLDNIREQVEEEAEGKADLQRQLSKANAEAQLWRSKYESEGVARSEELEEAKRKLQARLAEAEETIESLNQKVVALEKTKQRLATEVEDLQLEVDRATAIANAAEKKQKAFDKIIGEWKLKVDDLAAELDASQKECRNYSTELFRLKGAYEEGQEQLEAVRRENKNLADEVKDLLDQIGEGGRNIHEIEKARKRLEAEKDELQAALEEAEAALEQEENKVLRAQLELSQVRQEIDRRIQEKEEEFENTRKNHQRALDSMQASLEAEAKGKAEALRMKKKLEADINELEIALDHANKANAEAQKNIKRYQAQIKDLQTALEEEQRARDDAREQLGISERRANALQNELEESRTLLEQADRARRQAEQELSDAHEQLNELSAQSASLSAAKRKLESELQTLHSDLDELLNEAKNSEEKAKKAMVDAARLADELRAEQDHAQTQEKLRKALEQQIKELQVRLDEAEANALKGGKKAIQKLEQRVRELENELDGEQRRHADAQKNLRKSERRIKELTFQAEEDRKNHERMQDLVDKLQQKIKTYKRQIEEAEEIAALNLAKFRKAQQELEEAEERADLAEQAISKFRGKGRAGSAARGVSPAPQRSRPALADGFGTFPPRFDLAPEDF
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Uniprot
A0A2W1BNN9
B2DBI1
A0A3S2NAJ9
A0A1E1WSK3
A0A2J7QRJ5
A0A2J7QRJ3
+ More
A0A2J7QRK4 A0A2J7QRK5 A0A2J7QRI1 A0A1P8BK10 A0A2J7QRJ1 A0A1P8BK01 A0A1B1NJA9 A0A151J2G8 A0A1P8BK26 A0A1B1NJB2 A0A2J7QRI4 A0A1B1NJ73 A0A2J7QRI3 A0A2J7QRK8 A0A1L8E5A3 A0A1P8BJZ9 A0A1L8E5E8 A0A1B1NJ54 A0A3L8DA84 A0A1P8BK09 A0A2J7QRK6 A0A195CR64 A0A1P8BK05 A0A1B1NJ80 A0A026W340 A0A1P8BK27 A0A1P8BK00 A0A195AV24 A0A1B1NJ69 A0A195FLL5 A0A1B1NJ71 A0A1B1NJ55 A0A1P8BK03 A0A2J7QRH6 A0A1B1NJA0 A0A1P8BJZ8 A0A1D1XV65 A0A1P8BJZ5 A0A1P8BJZ7 A0A1B1NJ57 A0A1S4H338 A0A1P8BJZ4 A0A1B1NJ58 K7IRT4 E0VCW3 A0A1B1NJ99 A0A1B1NJ74 A0A1S4H419 A0A1S4H3X2 A0A1S4H333 A0A1S4H334 A0A1L8E5H7 A0A1S4H350 A0A1W4WPY6 A0A2M4CTP0 A0A1W4WDS2 A0A1S4H329 A0A1S4H414 A0A1W4WPS8 A0A1W4WPS3 A0A1W4WPR2 A0A1D1Z8S6 A0A1W4WQ10 A0A1W4WDQ7 A0A1W4WPU1 A0A1W4WDW1 A0A1S4H3W7 A0A1W4WNR4 A0A1W4WDU4 A0A2M4AEE0 A0A1W4WDX5 A0A1W4WPR7 A0A1W4WDT9 A0A1W4WPU4 A0A1L8E579 A0A1W4WNQ4 A0A1W4WDT2 A0A1W4WQ15 A0A1Q3G4T4 A0A1S4H355 A0A2M4CTJ8 A0A1W4WDU7 A0A1W4WDU9 A0A1W4WDR6 A0A1W4WDT7 A0A1W4WDW5 A0A1S4H339 A0A1W4WNR8 A0A1W4WDX0 A0A1I8M404 A0A1W4WPT2 A0A1I8M420 A0A1W4WDR2 A0A0Q9X403
A0A2J7QRK4 A0A2J7QRK5 A0A2J7QRI1 A0A1P8BK10 A0A2J7QRJ1 A0A1P8BK01 A0A1B1NJA9 A0A151J2G8 A0A1P8BK26 A0A1B1NJB2 A0A2J7QRI4 A0A1B1NJ73 A0A2J7QRI3 A0A2J7QRK8 A0A1L8E5A3 A0A1P8BJZ9 A0A1L8E5E8 A0A1B1NJ54 A0A3L8DA84 A0A1P8BK09 A0A2J7QRK6 A0A195CR64 A0A1P8BK05 A0A1B1NJ80 A0A026W340 A0A1P8BK27 A0A1P8BK00 A0A195AV24 A0A1B1NJ69 A0A195FLL5 A0A1B1NJ71 A0A1B1NJ55 A0A1P8BK03 A0A2J7QRH6 A0A1B1NJA0 A0A1P8BJZ8 A0A1D1XV65 A0A1P8BJZ5 A0A1P8BJZ7 A0A1B1NJ57 A0A1S4H338 A0A1P8BJZ4 A0A1B1NJ58 K7IRT4 E0VCW3 A0A1B1NJ99 A0A1B1NJ74 A0A1S4H419 A0A1S4H3X2 A0A1S4H333 A0A1S4H334 A0A1L8E5H7 A0A1S4H350 A0A1W4WPY6 A0A2M4CTP0 A0A1W4WDS2 A0A1S4H329 A0A1S4H414 A0A1W4WPS8 A0A1W4WPS3 A0A1W4WPR2 A0A1D1Z8S6 A0A1W4WQ10 A0A1W4WDQ7 A0A1W4WPU1 A0A1W4WDW1 A0A1S4H3W7 A0A1W4WNR4 A0A1W4WDU4 A0A2M4AEE0 A0A1W4WDX5 A0A1W4WPR7 A0A1W4WDT9 A0A1W4WPU4 A0A1L8E579 A0A1W4WNQ4 A0A1W4WDT2 A0A1W4WQ15 A0A1Q3G4T4 A0A1S4H355 A0A2M4CTJ8 A0A1W4WDU7 A0A1W4WDU9 A0A1W4WDR6 A0A1W4WDT7 A0A1W4WDW5 A0A1S4H339 A0A1W4WNR8 A0A1W4WDX0 A0A1I8M404 A0A1W4WPT2 A0A1I8M420 A0A1W4WDR2 A0A0Q9X403
EMBL
KZ150021
PZC74897.1
AB264661
BAG30740.1
RSAL01000147
RVE45896.1
+ More
GDQN01001263 JAT89791.1 NEVH01011894 PNF31196.1 PNF31209.1 PNF31203.1 PNF31205.1 PNF31201.1 KU516821 ANS83653.1 PNF31208.1 ANS83654.1 KU533788 ANS83723.1 KQ980396 KYN16227.1 ANS83651.1 KU533789 ANS83724.1 PNF31204.1 KU533786 ANS83721.1 PNF31198.1 PNF31207.1 GFDF01000157 JAV13927.1 ANS83650.1 GFDF01000156 JAV13928.1 KU533785 ANS83720.1 QOIP01000010 RLU17397.1 ANS83656.1 PNF31206.1 KQ977381 KYN03196.1 ANS83657.1 KU533791 ANS83726.1 KK107455 EZA50495.1 ANS83652.1 ANS83649.1 KQ976736 KYM76093.1 KU533792 ANS83727.1 KQ981490 KYN41308.1 KU533787 ANS83722.1 KU533784 ANS83719.1 ANS83655.1 PNF31202.1 KU533790 ANS83725.1 ANS83647.1 GDJX01021654 JAT46282.1 ANS83648.1 ANS83646.1 KU533782 ANS83717.1 AAAB01008980 ANS83645.1 KU533783 ANS83718.1 DS235065 EEB11219.1 KU533780 ANS83715.1 KU533781 ANS83716.1 GFDF01000158 JAV13926.1 GGFL01004461 MBW68639.1 GDJX01004640 JAT63296.1 GGFK01005834 MBW39155.1 GFDF01000184 JAV13900.1 GFDL01000266 JAV34779.1 GGFL01004462 MBW68640.1 CH933807 KRG02605.1
GDQN01001263 JAT89791.1 NEVH01011894 PNF31196.1 PNF31209.1 PNF31203.1 PNF31205.1 PNF31201.1 KU516821 ANS83653.1 PNF31208.1 ANS83654.1 KU533788 ANS83723.1 KQ980396 KYN16227.1 ANS83651.1 KU533789 ANS83724.1 PNF31204.1 KU533786 ANS83721.1 PNF31198.1 PNF31207.1 GFDF01000157 JAV13927.1 ANS83650.1 GFDF01000156 JAV13928.1 KU533785 ANS83720.1 QOIP01000010 RLU17397.1 ANS83656.1 PNF31206.1 KQ977381 KYN03196.1 ANS83657.1 KU533791 ANS83726.1 KK107455 EZA50495.1 ANS83652.1 ANS83649.1 KQ976736 KYM76093.1 KU533792 ANS83727.1 KQ981490 KYN41308.1 KU533787 ANS83722.1 KU533784 ANS83719.1 ANS83655.1 PNF31202.1 KU533790 ANS83725.1 ANS83647.1 GDJX01021654 JAT46282.1 ANS83648.1 ANS83646.1 KU533782 ANS83717.1 AAAB01008980 ANS83645.1 KU533783 ANS83718.1 DS235065 EEB11219.1 KU533780 ANS83715.1 KU533781 ANS83716.1 GFDF01000158 JAV13926.1 GGFL01004461 MBW68639.1 GDJX01004640 JAT63296.1 GGFK01005834 MBW39155.1 GFDF01000184 JAV13900.1 GFDL01000266 JAV34779.1 GGFL01004462 MBW68640.1 CH933807 KRG02605.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
A0A2W1BNN9
B2DBI1
A0A3S2NAJ9
A0A1E1WSK3
A0A2J7QRJ5
A0A2J7QRJ3
+ More
A0A2J7QRK4 A0A2J7QRK5 A0A2J7QRI1 A0A1P8BK10 A0A2J7QRJ1 A0A1P8BK01 A0A1B1NJA9 A0A151J2G8 A0A1P8BK26 A0A1B1NJB2 A0A2J7QRI4 A0A1B1NJ73 A0A2J7QRI3 A0A2J7QRK8 A0A1L8E5A3 A0A1P8BJZ9 A0A1L8E5E8 A0A1B1NJ54 A0A3L8DA84 A0A1P8BK09 A0A2J7QRK6 A0A195CR64 A0A1P8BK05 A0A1B1NJ80 A0A026W340 A0A1P8BK27 A0A1P8BK00 A0A195AV24 A0A1B1NJ69 A0A195FLL5 A0A1B1NJ71 A0A1B1NJ55 A0A1P8BK03 A0A2J7QRH6 A0A1B1NJA0 A0A1P8BJZ8 A0A1D1XV65 A0A1P8BJZ5 A0A1P8BJZ7 A0A1B1NJ57 A0A1S4H338 A0A1P8BJZ4 A0A1B1NJ58 K7IRT4 E0VCW3 A0A1B1NJ99 A0A1B1NJ74 A0A1S4H419 A0A1S4H3X2 A0A1S4H333 A0A1S4H334 A0A1L8E5H7 A0A1S4H350 A0A1W4WPY6 A0A2M4CTP0 A0A1W4WDS2 A0A1S4H329 A0A1S4H414 A0A1W4WPS8 A0A1W4WPS3 A0A1W4WPR2 A0A1D1Z8S6 A0A1W4WQ10 A0A1W4WDQ7 A0A1W4WPU1 A0A1W4WDW1 A0A1S4H3W7 A0A1W4WNR4 A0A1W4WDU4 A0A2M4AEE0 A0A1W4WDX5 A0A1W4WPR7 A0A1W4WDT9 A0A1W4WPU4 A0A1L8E579 A0A1W4WNQ4 A0A1W4WDT2 A0A1W4WQ15 A0A1Q3G4T4 A0A1S4H355 A0A2M4CTJ8 A0A1W4WDU7 A0A1W4WDU9 A0A1W4WDR6 A0A1W4WDT7 A0A1W4WDW5 A0A1S4H339 A0A1W4WNR8 A0A1W4WDX0 A0A1I8M404 A0A1W4WPT2 A0A1I8M420 A0A1W4WDR2 A0A0Q9X403
A0A2J7QRK4 A0A2J7QRK5 A0A2J7QRI1 A0A1P8BK10 A0A2J7QRJ1 A0A1P8BK01 A0A1B1NJA9 A0A151J2G8 A0A1P8BK26 A0A1B1NJB2 A0A2J7QRI4 A0A1B1NJ73 A0A2J7QRI3 A0A2J7QRK8 A0A1L8E5A3 A0A1P8BJZ9 A0A1L8E5E8 A0A1B1NJ54 A0A3L8DA84 A0A1P8BK09 A0A2J7QRK6 A0A195CR64 A0A1P8BK05 A0A1B1NJ80 A0A026W340 A0A1P8BK27 A0A1P8BK00 A0A195AV24 A0A1B1NJ69 A0A195FLL5 A0A1B1NJ71 A0A1B1NJ55 A0A1P8BK03 A0A2J7QRH6 A0A1B1NJA0 A0A1P8BJZ8 A0A1D1XV65 A0A1P8BJZ5 A0A1P8BJZ7 A0A1B1NJ57 A0A1S4H338 A0A1P8BJZ4 A0A1B1NJ58 K7IRT4 E0VCW3 A0A1B1NJ99 A0A1B1NJ74 A0A1S4H419 A0A1S4H3X2 A0A1S4H333 A0A1S4H334 A0A1L8E5H7 A0A1S4H350 A0A1W4WPY6 A0A2M4CTP0 A0A1W4WDS2 A0A1S4H329 A0A1S4H414 A0A1W4WPS8 A0A1W4WPS3 A0A1W4WPR2 A0A1D1Z8S6 A0A1W4WQ10 A0A1W4WDQ7 A0A1W4WPU1 A0A1W4WDW1 A0A1S4H3W7 A0A1W4WNR4 A0A1W4WDU4 A0A2M4AEE0 A0A1W4WDX5 A0A1W4WPR7 A0A1W4WDT9 A0A1W4WPU4 A0A1L8E579 A0A1W4WNQ4 A0A1W4WDT2 A0A1W4WQ15 A0A1Q3G4T4 A0A1S4H355 A0A2M4CTJ8 A0A1W4WDU7 A0A1W4WDU9 A0A1W4WDR6 A0A1W4WDT7 A0A1W4WDW5 A0A1S4H339 A0A1W4WNR8 A0A1W4WDX0 A0A1I8M404 A0A1W4WPT2 A0A1I8M420 A0A1W4WDR2 A0A0Q9X403
PDB
3JBH
E-value=0,
Score=5496
Ontologies
GO
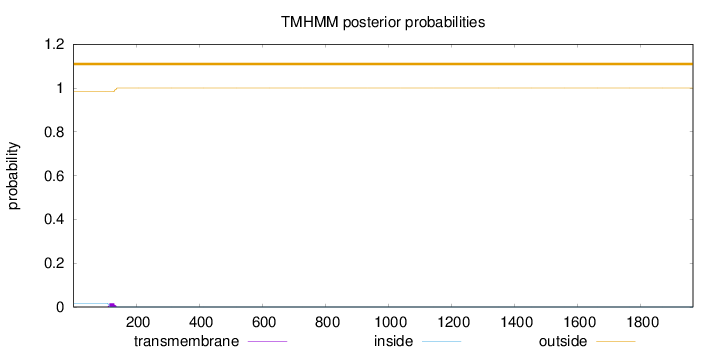
Topology
Length:
1967
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.340520000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01680
outside
1 - 1967
Population Genetic Test Statistics
Pi
185.188255
Theta
199.021831
Tajima's D
-0.700366
CLR
1.487052
CSRT
0.189940502974851
Interpretation
Uncertain
