Gene
KWMTBOMO04265
Annotation
PREDICTED:_E3_ubiquitin-protein_ligase_UBR3-like_[Papilio_xuthus]
Full name
E3 ubiquitin-protein ligase Ubr3
Alternative Name
E3 ubiquitin-protein transferase Ubr3
Location in the cell
PlasmaMembrane Reliability : 1.914
Sequence
CDS
ATGGCTAATTCAACGGCTCAAGTGTTAATGAAGAAGGGTAAAAGGGGGGCAGCCGCTTACATACATGCGGACTGCTCCAGCGGATCACCACAGCACTTGGGGCCACTTCTAGACGTTCTGCTGAACCCTAGCAAGGCTATAGACGAGTGGGAGACCATTGACTGGTGTCGGTGGCTGCTGGCTGGAGGTAGAACACCAGATGAATTCGCTGCAGTCGTGCGAAGCTACGACAAACATGACAAGTGCGGCCTAGTTTGGATACCGCGCGTGGTCGCCTACCGCTGTCGGACGTGCGGTATATCGCCCTGCATGTCGATATGCAGAGAATGCTTCCACCGAGGCGATCACTCCACGCACGACTTCAACATGTTTCTATCGCAGGCCGGTGGCGCTTGCGACTGTGGTGATAAGTCCGTCATGAAGGAAGACGGATTCTGCAGTAACCACGGTAATAAATGTCCTCGGCCCGGCACCGTTCCCGCGGAACTGATGTGTGTTGCTGAAGCGATGATGCCGCGGCTCATACTACGGCTGCTCCAGCACTTCAGGGAGAACAGCAGCGGAAGCCAAACCAATTCAGACCGGTGTCGTATTGCGGTTCAAGAGTGCGAGGGCTACGTTCAGATGTTGATGGAGTTCAATAACATGGGTGACCTCATGCGGTCGGCCATGACTAAGGCGCTTATTAACCCACAGATGTACAGGAATCTTGTGGTTCCACCATTCCCGGACACGGAGTACGGGGCGTACATGGCAGAGTCACACCACATGTACGAGAGGGCGTTAGATCTGTTCCCAGCACCGGAGCCGCCTGACGAGTACTGCCACCTGCCGGCGCTCGCGCCTCGACTGACGCACAACACGCTGTTGGAAGAATTCATATTCTGGACTTTCAAATACGAGTTTCCGCAGAACGTGGTCTGCTTCTTGCTGAATATGTTACCCGACCAGGACTACAAGGAGCACCTGACGCGCACGTTCGTGCTGCACTACGGGCGCATCCCGCTGGTGCTGGAGGCGGCCGCCGACCCCGACACGCTGTCCAACCGCGTCGTGCACATGTCCGTGCAGCTGTTCTCCAACGAGGCGCTGGCGCTGCGCTGCGTGCAGCACCTGCACCTGCTGCACGTCATGGTGCTCTCGCTGCGCCTCATGATGGCCAAGATCCTCGTCCAGAACACGCTGCACGACCCCCGCCGCAACTACCACCGCGTCATCGACTGCACGCAGAAGGTCATGAAGGAGCACTGCTACTGGCCGCTCGTGTCCGACTTCAACAACGTGCTCTCGCACAAGAGCGTCGCGCTGCTGTTCCTGCAGGACGACGCGCTCATCGACATGTGGTTCGAGTTCCTGTCCATGCTGCAAGGCATGAACGTGAACGTGCGCGAGACCGGCGGGCACATCGAGTTCGAGCCGTCGTCGTACTACGCGGCGTTCTCGTGCGAGCTGGAGGCGGCCGCGTACCCCATGTGGTCCGTGCTGTCGCACCTGACGGCGCCCGAGCACGCCCCGCTGGCGCGCCGCATGCTGGGCGCGGCGCTGGGCAGCCTGCGCGCGTGGCTGGGCGCCGTGCAGTGCACCGAGCTGCGCCCCGAGCGCGACGCCGCCATGCGCGCCTCCTTCCACTTCCCGCTGCACCGCTACCTCGCCGCCTTCCTGTGCGCCGCCGTGCGCTGGATGGGCGTGCGCGCCGCCGACGTGCTGCCGCCCGCGCCGCTGCTCGCGCTGCTGGCCGCGCACCCGCTGCGCGTGCAGGTGTGA
Protein
MANSTAQVLMKKGKRGAAAYIHADCSSGSPQHLGPLLDVLLNPSKAIDEWETIDWCRWLLAGGRTPDEFAAVVRSYDKHDKCGLVWIPRVVAYRCRTCGISPCMSICRECFHRGDHSTHDFNMFLSQAGGACDCGDKSVMKEDGFCSNHGNKCPRPGTVPAELMCVAEAMMPRLILRLLQHFRENSSGSQTNSDRCRIAVQECEGYVQMLMEFNNMGDLMRSAMTKALINPQMYRNLVVPPFPDTEYGAYMAESHHMYERALDLFPAPEPPDEYCHLPALAPRLTHNTLLEEFIFWTFKYEFPQNVVCFLLNMLPDQDYKEHLTRTFVLHYGRIPLVLEAAADPDTLSNRVVHMSVQLFSNEALALRCVQHLHLLHVMVLSLRLMMAKILVQNTLHDPRRNYHRVIDCTQKVMKEHCYWPLVSDFNNVLSHKSVALLFLQDDALIDMWFEFLSMLQGMNVNVRETGGHIEFEPSSYYAAFSCELEAAAYPMWSVLSHLTAPEHAPLARRMLGAALGSLRAWLGAVQCTELRPERDAAMRASFHFPLHRYLAAFLCAAVRWMGVRAADVLPPAPLLALLAAHPLRVQV
Summary
Description
E3 ubiquitin-protein ligase which is a component of the N-end rule pathway (PubMed:26383956, PubMed:27195754). Recognizes and binds to proteins bearing specific N-terminal residues, leading to their ubiquitination and subsequent degradation (PubMed:26383956, PubMed:27195754). Binds to the E3 ubiquitin-protein ligase Diap1 and enhances its ubiquitination and anti-apoptotic functions (PubMed:25146930). Essential during trichome development for the ubiquitination of the N-terminus of ovo isoform B (svb), converting it from a transcriptional inhibitor to an activator (PubMed:26383956). Positively regulates a hh-signaling pathway which functions in photoreceptor differentiation (PubMed:27195754). Activation of hh up-regulates transcription of Ubr3, which in turn promotes hh signaling by mediating the ubiquitination and degradation of cos (PubMed:27195754). Necessary for auditory transduction: plays a role in Johnston's organ organization by acting in the regulation of zip and ck function in scolopidial apical attachment (PubMed:27331610). Likely to function by acting in a pathway that negatively regulates the ubiquitination of zip, consequently affecting its interaction with ck (PubMed:27331610). May also negatively regulate a component of the SCF (SKP1-CUL1-F-box protein) E3 ubiquitin-protein ligase complex Cul1, which also appears to function in the negative regulation of the zip-ck interaction and scolopidial apical attachment (PubMed:27331610).
Catalytic Activity
S-ubiquitinyl-[E2 ubiquitin-conjugating enzyme]-L-cysteine + [acceptor protein]-L-lysine = [E2 ubiquitin-conjugating enzyme]-L-cysteine + N(6)-ubiquitinyl-[acceptor protein]-L-lysine.
Subunit
Selectively interacts (via UBR-type zinc finger) with the cleaved form of Diap1; this interaction is enhanced by tal (PubMed:25146930, PubMed:26383956). Interacts with tal and Rrp1 (PubMed:26383956). Interacts with ovo isoform B (via N-terminus) (PubMed:26383956). Interacts with Cad99C (via the cytoplasmic domain) (PubMed:27331610). Interacts with ck and Sans (PubMed:27331610). Interacts with cos (via Kinesin motor domain) (PubMed:27195754).
Similarity
Belongs to the UBR1 family.
Keywords
Alternative splicing
Complete proteome
Cytoplasm
Metal-binding
Nucleus
Reference proteome
Transferase
Ubl conjugation
Ubl conjugation pathway
Zinc
Zinc-finger
Feature
chain E3 ubiquitin-protein ligase Ubr3
splice variant In isoform C.
splice variant In isoform C.
Uniprot
A0A2H1VFJ5
A0A3S2NVT3
A0A0N1PGV0
A0A212EZ46
A0A2A4IYP5
A0A1Q3EZQ2
+ More
A0A1Q3EZK2 A0A1S4G7C6 A0A1Y9J0I4 A0A1I8JT39 A0A034VWB8 W8B5F0 A0A0K8UIB1 A0A182KJG6 Q7QFA9 A0A0A1XH15 A0A0K8VKQ6 A0A182V1V5 A0A182Q4Q3 A0A182YCW1 A0A182M9J8 W5JUH5 A0A1Y1LL35 B3NY00 A0A1W4VIB7 A0A1W4VV28 M9PDZ8 B4Q0P6 Q9W3M3-2 A0A1A9YI71 Q9W3M3 B4L539 B4IL59 A0A1B0AHV8 A0A1B0GEG5 B3MRI6 B4M712 A0A182FJ44 B4JLX5 B5DKT1 A0A1I8MYG3 B4H3V1 B4N1W8 A0A3B0JS14 A0A1I8PE15 E2B3J3 A0A0M4ERC5 A0A1B0D3K3 A0A0T6B1C4 A0A0J7L456 A0A195C4T0 A0A195FM35 A0A3L8E1S8 A0A0K8VTC5 A0A195AVD5 F4WEJ4 A0A139WE03 A0A2J7PBM3 A0A182GR11 E1ZZB8 A0A232FHL6 B0WQY2 A0A1B6EPK5 A0A2M4ALH5 A0A1B6L2X0 A0A0L0C251 A0A182N8H4 A0A1B6CS83 A0A1B6CK60 A0A026WLR4 A0A1B6J6P5 A0A154PCP2 A0A182PR12 A0A182W5P2 A0A088ACJ7 A0A182SJ28 A0A023EZG5 A0A1A9UZP9 A0A1B0B2C7 A0A0C9QRP8 A0A069DXT5 A0A151X868 A0A0C9RDL4 A0A3Q0ISP9 Q16LM4 A0A0A9Y5B1 A0A336M7M7 A0A0A9XD93 A0A0L7QYZ4 A0A2P8YCV6 A0A2A3EQ11 A0A0P5NH07 A0A0P5X2M7 A0A0P5K5Z8 A0A0P6ALY1 A0A0P5DVB8 A0A0P6A1A3 A0A0P5LMZ2
A0A1Q3EZK2 A0A1S4G7C6 A0A1Y9J0I4 A0A1I8JT39 A0A034VWB8 W8B5F0 A0A0K8UIB1 A0A182KJG6 Q7QFA9 A0A0A1XH15 A0A0K8VKQ6 A0A182V1V5 A0A182Q4Q3 A0A182YCW1 A0A182M9J8 W5JUH5 A0A1Y1LL35 B3NY00 A0A1W4VIB7 A0A1W4VV28 M9PDZ8 B4Q0P6 Q9W3M3-2 A0A1A9YI71 Q9W3M3 B4L539 B4IL59 A0A1B0AHV8 A0A1B0GEG5 B3MRI6 B4M712 A0A182FJ44 B4JLX5 B5DKT1 A0A1I8MYG3 B4H3V1 B4N1W8 A0A3B0JS14 A0A1I8PE15 E2B3J3 A0A0M4ERC5 A0A1B0D3K3 A0A0T6B1C4 A0A0J7L456 A0A195C4T0 A0A195FM35 A0A3L8E1S8 A0A0K8VTC5 A0A195AVD5 F4WEJ4 A0A139WE03 A0A2J7PBM3 A0A182GR11 E1ZZB8 A0A232FHL6 B0WQY2 A0A1B6EPK5 A0A2M4ALH5 A0A1B6L2X0 A0A0L0C251 A0A182N8H4 A0A1B6CS83 A0A1B6CK60 A0A026WLR4 A0A1B6J6P5 A0A154PCP2 A0A182PR12 A0A182W5P2 A0A088ACJ7 A0A182SJ28 A0A023EZG5 A0A1A9UZP9 A0A1B0B2C7 A0A0C9QRP8 A0A069DXT5 A0A151X868 A0A0C9RDL4 A0A3Q0ISP9 Q16LM4 A0A0A9Y5B1 A0A336M7M7 A0A0A9XD93 A0A0L7QYZ4 A0A2P8YCV6 A0A2A3EQ11 A0A0P5NH07 A0A0P5X2M7 A0A0P5K5Z8 A0A0P6ALY1 A0A0P5DVB8 A0A0P6A1A3 A0A0P5LMZ2
EC Number
2.3.2.27
Pubmed
26354079
22118469
17510324
25348373
24495485
20966253
+ More
12364791 25830018 25244985 20920257 23761445 28004739 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 25146930 26383956 27331610 27195754 15632085 25315136 20798317 30249741 21719571 18362917 19820115 26483478 28648823 26108605 24508170 25474469 26334808 25401762 29403074
12364791 25830018 25244985 20920257 23761445 28004739 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 25146930 26383956 27331610 27195754 15632085 25315136 20798317 30249741 21719571 18362917 19820115 26483478 28648823 26108605 24508170 25474469 26334808 25401762 29403074
EMBL
ODYU01002297
SOQ39599.1
RSAL01000147
RVE45887.1
KQ460480
KPJ14353.1
+ More
AGBW02011366 OWR46775.1 NWSH01004918 PCG64586.1 GFDL01014260 JAV20785.1 GFDL01014321 JAV20724.1 APCN01001350 APCN01001351 APCN01001352 APCN01001353 APCN01001354 GAKP01012817 JAC46135.1 GAMC01018044 JAB88511.1 GDHF01025872 JAI26442.1 AAAB01008846 EAA06288.4 GBXI01004051 JAD10241.1 GDHF01012847 JAI39467.1 AXCN02002232 AXCM01001731 AXCM01001732 ADMH02000109 ETN67801.1 GEZM01054581 JAV73548.1 CH954180 EDV47521.2 AE014298 AGB95162.1 CM000162 EDX02317.2 CH933811 EDW06298.2 CH480865 EDW53676.1 CCAG010012254 CH902622 EDV34391.2 CH940653 EDW62579.2 CH916371 EDV91736.1 CH379063 EDY72131.2 CH479207 EDW31052.1 CH963925 EDW78357.2 OUUW01000003 SPP78270.1 GL445346 EFN89745.1 CP012528 ALC48652.1 AJVK01002934 AJVK01002935 LJIG01016242 KRT81190.1 LBMM01000713 KMQ97672.1 KQ978287 KYM95615.1 KQ981490 KYN41386.1 QOIP01000001 RLU26482.1 GDHF01010529 JAI41785.1 KQ976736 KYM76022.1 GL888103 EGI67357.1 KQ971357 KYB26142.1 NEVH01027081 PNF13728.1 JXUM01016250 JXUM01016251 KQ560453 KXJ82342.1 GL435311 EFN73533.1 NNAY01000179 OXU30256.1 DS232047 EDS33069.1 GECZ01029892 JAS39877.1 GGFK01008276 MBW41597.1 GEBQ01021986 JAT17991.1 JRES01000987 KNC26385.1 GEDC01021027 JAS16271.1 GEDC01023496 JAS13802.1 KK107154 EZA56898.1 GECU01012874 JAS94832.1 KQ434874 KZC09699.1 GBBI01004070 JAC14642.1 JXJN01007607 GBYB01006394 JAG76161.1 GBGD01000051 JAC88838.1 KQ982430 KYQ56571.1 GBYB01006395 JAG76162.1 CH477900 EAT35223.1 GBHO01017316 JAG26288.1 UFQT01000666 SSX26322.1 GBHO01025635 JAG17969.1 KQ414685 KOC63813.1 PYGN01000697 PSN42091.1 KZ288204 PBC33219.1 GDIQ01142504 JAL09222.1 GDIP01078324 JAM25391.1 GDIQ01188690 JAK63035.1 GDIP01040978 JAM62737.1 GDIP01152167 JAJ71235.1 GDIP01036932 JAM66783.1 GDIQ01169742 JAK81983.1
AGBW02011366 OWR46775.1 NWSH01004918 PCG64586.1 GFDL01014260 JAV20785.1 GFDL01014321 JAV20724.1 APCN01001350 APCN01001351 APCN01001352 APCN01001353 APCN01001354 GAKP01012817 JAC46135.1 GAMC01018044 JAB88511.1 GDHF01025872 JAI26442.1 AAAB01008846 EAA06288.4 GBXI01004051 JAD10241.1 GDHF01012847 JAI39467.1 AXCN02002232 AXCM01001731 AXCM01001732 ADMH02000109 ETN67801.1 GEZM01054581 JAV73548.1 CH954180 EDV47521.2 AE014298 AGB95162.1 CM000162 EDX02317.2 CH933811 EDW06298.2 CH480865 EDW53676.1 CCAG010012254 CH902622 EDV34391.2 CH940653 EDW62579.2 CH916371 EDV91736.1 CH379063 EDY72131.2 CH479207 EDW31052.1 CH963925 EDW78357.2 OUUW01000003 SPP78270.1 GL445346 EFN89745.1 CP012528 ALC48652.1 AJVK01002934 AJVK01002935 LJIG01016242 KRT81190.1 LBMM01000713 KMQ97672.1 KQ978287 KYM95615.1 KQ981490 KYN41386.1 QOIP01000001 RLU26482.1 GDHF01010529 JAI41785.1 KQ976736 KYM76022.1 GL888103 EGI67357.1 KQ971357 KYB26142.1 NEVH01027081 PNF13728.1 JXUM01016250 JXUM01016251 KQ560453 KXJ82342.1 GL435311 EFN73533.1 NNAY01000179 OXU30256.1 DS232047 EDS33069.1 GECZ01029892 JAS39877.1 GGFK01008276 MBW41597.1 GEBQ01021986 JAT17991.1 JRES01000987 KNC26385.1 GEDC01021027 JAS16271.1 GEDC01023496 JAS13802.1 KK107154 EZA56898.1 GECU01012874 JAS94832.1 KQ434874 KZC09699.1 GBBI01004070 JAC14642.1 JXJN01007607 GBYB01006394 JAG76161.1 GBGD01000051 JAC88838.1 KQ982430 KYQ56571.1 GBYB01006395 JAG76162.1 CH477900 EAT35223.1 GBHO01017316 JAG26288.1 UFQT01000666 SSX26322.1 GBHO01025635 JAG17969.1 KQ414685 KOC63813.1 PYGN01000697 PSN42091.1 KZ288204 PBC33219.1 GDIQ01142504 JAL09222.1 GDIP01078324 JAM25391.1 GDIQ01188690 JAK63035.1 GDIP01040978 JAM62737.1 GDIP01152167 JAJ71235.1 GDIP01036932 JAM66783.1 GDIQ01169742 JAK81983.1
Proteomes
UP000283053
UP000053240
UP000007151
UP000218220
UP000076407
UP000075840
+ More
UP000075882 UP000007062 UP000075903 UP000075886 UP000076408 UP000075883 UP000000673 UP000008711 UP000192221 UP000000803 UP000002282 UP000092443 UP000009192 UP000001292 UP000092445 UP000092444 UP000007801 UP000008792 UP000069272 UP000001070 UP000001819 UP000095301 UP000008744 UP000007798 UP000268350 UP000095300 UP000008237 UP000092553 UP000092462 UP000036403 UP000078542 UP000078541 UP000279307 UP000078540 UP000007755 UP000007266 UP000235965 UP000069940 UP000249989 UP000000311 UP000215335 UP000002320 UP000037069 UP000075884 UP000053097 UP000076502 UP000075885 UP000075920 UP000005203 UP000075901 UP000078200 UP000092460 UP000075809 UP000079169 UP000008820 UP000053825 UP000245037 UP000242457
UP000075882 UP000007062 UP000075903 UP000075886 UP000076408 UP000075883 UP000000673 UP000008711 UP000192221 UP000000803 UP000002282 UP000092443 UP000009192 UP000001292 UP000092445 UP000092444 UP000007801 UP000008792 UP000069272 UP000001070 UP000001819 UP000095301 UP000008744 UP000007798 UP000268350 UP000095300 UP000008237 UP000092553 UP000092462 UP000036403 UP000078542 UP000078541 UP000279307 UP000078540 UP000007755 UP000007266 UP000235965 UP000069940 UP000249989 UP000000311 UP000215335 UP000002320 UP000037069 UP000075884 UP000053097 UP000076502 UP000075885 UP000075920 UP000005203 UP000075901 UP000078200 UP000092460 UP000075809 UP000079169 UP000008820 UP000053825 UP000245037 UP000242457
Interpro
ProteinModelPortal
A0A2H1VFJ5
A0A3S2NVT3
A0A0N1PGV0
A0A212EZ46
A0A2A4IYP5
A0A1Q3EZQ2
+ More
A0A1Q3EZK2 A0A1S4G7C6 A0A1Y9J0I4 A0A1I8JT39 A0A034VWB8 W8B5F0 A0A0K8UIB1 A0A182KJG6 Q7QFA9 A0A0A1XH15 A0A0K8VKQ6 A0A182V1V5 A0A182Q4Q3 A0A182YCW1 A0A182M9J8 W5JUH5 A0A1Y1LL35 B3NY00 A0A1W4VIB7 A0A1W4VV28 M9PDZ8 B4Q0P6 Q9W3M3-2 A0A1A9YI71 Q9W3M3 B4L539 B4IL59 A0A1B0AHV8 A0A1B0GEG5 B3MRI6 B4M712 A0A182FJ44 B4JLX5 B5DKT1 A0A1I8MYG3 B4H3V1 B4N1W8 A0A3B0JS14 A0A1I8PE15 E2B3J3 A0A0M4ERC5 A0A1B0D3K3 A0A0T6B1C4 A0A0J7L456 A0A195C4T0 A0A195FM35 A0A3L8E1S8 A0A0K8VTC5 A0A195AVD5 F4WEJ4 A0A139WE03 A0A2J7PBM3 A0A182GR11 E1ZZB8 A0A232FHL6 B0WQY2 A0A1B6EPK5 A0A2M4ALH5 A0A1B6L2X0 A0A0L0C251 A0A182N8H4 A0A1B6CS83 A0A1B6CK60 A0A026WLR4 A0A1B6J6P5 A0A154PCP2 A0A182PR12 A0A182W5P2 A0A088ACJ7 A0A182SJ28 A0A023EZG5 A0A1A9UZP9 A0A1B0B2C7 A0A0C9QRP8 A0A069DXT5 A0A151X868 A0A0C9RDL4 A0A3Q0ISP9 Q16LM4 A0A0A9Y5B1 A0A336M7M7 A0A0A9XD93 A0A0L7QYZ4 A0A2P8YCV6 A0A2A3EQ11 A0A0P5NH07 A0A0P5X2M7 A0A0P5K5Z8 A0A0P6ALY1 A0A0P5DVB8 A0A0P6A1A3 A0A0P5LMZ2
A0A1Q3EZK2 A0A1S4G7C6 A0A1Y9J0I4 A0A1I8JT39 A0A034VWB8 W8B5F0 A0A0K8UIB1 A0A182KJG6 Q7QFA9 A0A0A1XH15 A0A0K8VKQ6 A0A182V1V5 A0A182Q4Q3 A0A182YCW1 A0A182M9J8 W5JUH5 A0A1Y1LL35 B3NY00 A0A1W4VIB7 A0A1W4VV28 M9PDZ8 B4Q0P6 Q9W3M3-2 A0A1A9YI71 Q9W3M3 B4L539 B4IL59 A0A1B0AHV8 A0A1B0GEG5 B3MRI6 B4M712 A0A182FJ44 B4JLX5 B5DKT1 A0A1I8MYG3 B4H3V1 B4N1W8 A0A3B0JS14 A0A1I8PE15 E2B3J3 A0A0M4ERC5 A0A1B0D3K3 A0A0T6B1C4 A0A0J7L456 A0A195C4T0 A0A195FM35 A0A3L8E1S8 A0A0K8VTC5 A0A195AVD5 F4WEJ4 A0A139WE03 A0A2J7PBM3 A0A182GR11 E1ZZB8 A0A232FHL6 B0WQY2 A0A1B6EPK5 A0A2M4ALH5 A0A1B6L2X0 A0A0L0C251 A0A182N8H4 A0A1B6CS83 A0A1B6CK60 A0A026WLR4 A0A1B6J6P5 A0A154PCP2 A0A182PR12 A0A182W5P2 A0A088ACJ7 A0A182SJ28 A0A023EZG5 A0A1A9UZP9 A0A1B0B2C7 A0A0C9QRP8 A0A069DXT5 A0A151X868 A0A0C9RDL4 A0A3Q0ISP9 Q16LM4 A0A0A9Y5B1 A0A336M7M7 A0A0A9XD93 A0A0L7QYZ4 A0A2P8YCV6 A0A2A3EQ11 A0A0P5NH07 A0A0P5X2M7 A0A0P5K5Z8 A0A0P6ALY1 A0A0P5DVB8 A0A0P6A1A3 A0A0P5LMZ2
PDB
3NY1
E-value=3.24666e-10,
Score=158
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
In Johnston's organ, high expression at the apical tips of neurons and scolopale cells (PubMed:27331610). In the cytoplasm expressed in puncta (PubMed:27195754). With evidence from 6 publications.
Nucleus In Johnston's organ, high expression at the apical tips of neurons and scolopale cells (PubMed:27331610). In the cytoplasm expressed in puncta (PubMed:27195754). With evidence from 6 publications.
Nucleus In Johnston's organ, high expression at the apical tips of neurons and scolopale cells (PubMed:27331610). In the cytoplasm expressed in puncta (PubMed:27195754). With evidence from 6 publications.
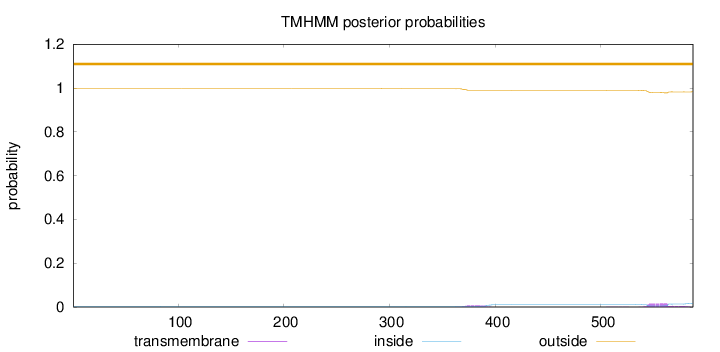
Length:
587
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.58543
Exp number, first 60 AAs:
0.0004
Total prob of N-in:
0.00288
outside
1 - 587
Population Genetic Test Statistics
Pi
191.855669
Theta
155.207085
Tajima's D
0.688067
CLR
0.444086
CSRT
0.570871456427179
Interpretation
Uncertain
