Pre Gene Modal
BGIBMGA014269
Annotation
PREDICTED:_uncharacterized_protein_LOC101735738_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.695
Sequence
CDS
ATGTCCGTTCGCTTCGTGCGCACGATTCTCGTGCTACTAGCGATCGTTCACAATTCGCGCGCTCTAGTCGATGAAGTCGTCGACGTACTCAAACTCGGGAAGGAGATAGGCGAAGAGATAATCGCATCATGGAACGTTGTAGGAAAGACGCTAAACGTATCGGAGGGCGTCGAACTCCCCCTAATCAGAAGACGTGAAAGATTAATACTGGCAAAACTCTCGCATATATCGCAATCGATCGACAGATTGGAGCTGGGAATAGAGAAGGCCGGTGCGGTGGCCTTGTTCCTGGCCAAGAACGGAGGGAGAGGAACCCGTTTTGAGTTGAAACTGCACGACATGACGGTCCTTCTGAACAAAGTGGCGGCAGTGGACAGACAGATGAGAGTGTACGTCGGCCTTCAAGAGGAATTGGAGCGAAGCACGTTGTTAGGCTTTGCTCAATCGTGTGTCTTTTACGAGCCGGATGCGCTACCTGGCGTGCTGGAGCAGATACACGCGCACATTGTTCCACCCCATAAGCTATTGTTAGGCAAGGGCCTTCTGCAGCAAATCGTCGAGGAAGTTCAGGTTGGTACAACACTGTATGAGAATTGTAACTTTGATACGTTATTATGA
Protein
MSVRFVRTILVLLAIVHNSRALVDEVVDVLKLGKEIGEEIIASWNVVGKTLNVSEGVELPLIRRRERLILAKLSHISQSIDRLELGIEKAGAVALFLAKNGGRGTRFELKLHDMTVLLNKVAAVDRQMRVYVGLQEELERSTLLGFAQSCVFYEPDALPGVLEQIHAHIVPPHKLLLGKGLLQQIVEEVQVGTTLYENCNFDTLL
Summary
Uniprot
H9JXK3
A0A2H1VGY2
A0A194Q182
A0A2W1BM01
A0A2W1BMQ5
A0A0N1IPA6
+ More
A0A0L7KTD6 S4PWW1 A0A212EZ97 A0A1E1WRF0 A0A1E1WM67 U4U0Y7 A0A336MDK9 D6WVE3 A0A336MGF5 A0A195FMC4 A0A0A1WEF3 A0A1B6ENR6 E1ZZB9 A0A1B0D3K5 A0A1B6D7N1 A0A1B6C4N6 W8BRC6 A0A1Y1NB04 A0A1J1I244 A0A0J7NZ76 A0A1A9WCI2 A0A0Q9X3L3 B3NXZ9 A0A158NF43 A0A0K8V3H6 A0A034WPQ5 A0A1A9YI70 A0A1B0B2D2 B4Q0P7 A0A1W6EW95 A0A195C615 A0A1B6IJS5 A0A1W4VV31 A0A0K8U3Z3 B3MRI5 Q9W3M4 A0A195E952 B4IL60 A0A2P8YCW1 T1PCL3 A0A1I8M2B6 B4M713 A0A1B0GEG6 B4L540 B0WQX7 A0A0M4ET40 A0A1B0ACS7 B4JLX4 A0A1A9UZP5 A0A067QHF3 A0A232FHW8 A0A1B0AHW0 A0A182GR10 A0A1I8PL78 A0A151X876 A0A1B0AHV6 Q29JA0 A0A0L0C4P0
A0A0L7KTD6 S4PWW1 A0A212EZ97 A0A1E1WRF0 A0A1E1WM67 U4U0Y7 A0A336MDK9 D6WVE3 A0A336MGF5 A0A195FMC4 A0A0A1WEF3 A0A1B6ENR6 E1ZZB9 A0A1B0D3K5 A0A1B6D7N1 A0A1B6C4N6 W8BRC6 A0A1Y1NB04 A0A1J1I244 A0A0J7NZ76 A0A1A9WCI2 A0A0Q9X3L3 B3NXZ9 A0A158NF43 A0A0K8V3H6 A0A034WPQ5 A0A1A9YI70 A0A1B0B2D2 B4Q0P7 A0A1W6EW95 A0A195C615 A0A1B6IJS5 A0A1W4VV31 A0A0K8U3Z3 B3MRI5 Q9W3M4 A0A195E952 B4IL60 A0A2P8YCW1 T1PCL3 A0A1I8M2B6 B4M713 A0A1B0GEG6 B4L540 B0WQX7 A0A0M4ET40 A0A1B0ACS7 B4JLX4 A0A1A9UZP5 A0A067QHF3 A0A232FHW8 A0A1B0AHW0 A0A182GR10 A0A1I8PL78 A0A151X876 A0A1B0AHV6 Q29JA0 A0A0L0C4P0
Pubmed
19121390
26354079
28756777
26227816
23622113
22118469
+ More
23537049 18362917 19820115 25830018 20798317 24495485 28004739 17994087 21347285 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 25315136 24845553 28648823 26483478 15632085 26108605
23537049 18362917 19820115 25830018 20798317 24495485 28004739 17994087 21347285 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 25315136 24845553 28648823 26483478 15632085 26108605
EMBL
BABH01032787
ODYU01002512
SOQ40105.1
KQ459580
KPI99342.1
KZ150080
+ More
PZC73896.1 KZ150021 PZC74905.1 KQ460480 KPJ14355.1 JTDY01005861 KOB66538.1 GAIX01006098 JAA86462.1 AGBW02011366 OWR46777.1 GDQN01001449 JAT89605.1 GDQN01003016 JAT88038.1 KB631763 ERL85943.1 UFQS01000919 UFQT01000919 SSX07672.1 SSX28010.1 KQ971357 EFA08553.2 SSX07671.1 SSX28009.1 KQ981490 KYN41387.1 GBXI01017201 JAC97090.1 GECZ01030212 JAS39557.1 GL435311 EFN73534.1 AJVK01002938 GEDC01015607 JAS21691.1 GEDC01029098 GEDC01001492 JAS08200.1 JAS35806.1 GAMC01007152 JAB99403.1 GEZM01013042 JAV92797.1 CVRI01000035 CRK92910.1 LBMM01000713 KMQ97670.1 CH963925 KRF98860.1 CH954180 EDV47520.2 ADTU01013733 GDHF01018860 JAI33454.1 GAKP01003249 JAC55703.1 JXJN01007607 CM000162 EDX02318.2 KY563580 ARK19989.1 KQ978287 KYM95616.1 GECU01020541 JAS87165.1 GDHF01030882 JAI21432.1 CH902622 EDV34390.2 AE014298 AY058541 AAF46301.1 AAL13770.1 KQ979479 KYN21359.1 CH480865 EDW53677.1 PYGN01000697 PSN42087.1 KA645693 AFP60322.1 CH940653 EDW62580.2 CCAG010012254 CH933811 EDW06299.2 DS232047 EDS33064.1 CP012528 ALC48651.1 CH916371 EDV91735.1 KK853387 KDR07959.1 NNAY01000179 OXU30262.1 JXUM01016245 JXUM01016246 JXUM01016247 JXUM01016248 JXUM01016249 KQ560453 KXJ82341.1 KQ982430 KYQ56572.1 CH379063 EAL32401.3 JRES01000987 KNC26404.1
PZC73896.1 KZ150021 PZC74905.1 KQ460480 KPJ14355.1 JTDY01005861 KOB66538.1 GAIX01006098 JAA86462.1 AGBW02011366 OWR46777.1 GDQN01001449 JAT89605.1 GDQN01003016 JAT88038.1 KB631763 ERL85943.1 UFQS01000919 UFQT01000919 SSX07672.1 SSX28010.1 KQ971357 EFA08553.2 SSX07671.1 SSX28009.1 KQ981490 KYN41387.1 GBXI01017201 JAC97090.1 GECZ01030212 JAS39557.1 GL435311 EFN73534.1 AJVK01002938 GEDC01015607 JAS21691.1 GEDC01029098 GEDC01001492 JAS08200.1 JAS35806.1 GAMC01007152 JAB99403.1 GEZM01013042 JAV92797.1 CVRI01000035 CRK92910.1 LBMM01000713 KMQ97670.1 CH963925 KRF98860.1 CH954180 EDV47520.2 ADTU01013733 GDHF01018860 JAI33454.1 GAKP01003249 JAC55703.1 JXJN01007607 CM000162 EDX02318.2 KY563580 ARK19989.1 KQ978287 KYM95616.1 GECU01020541 JAS87165.1 GDHF01030882 JAI21432.1 CH902622 EDV34390.2 AE014298 AY058541 AAF46301.1 AAL13770.1 KQ979479 KYN21359.1 CH480865 EDW53677.1 PYGN01000697 PSN42087.1 KA645693 AFP60322.1 CH940653 EDW62580.2 CCAG010012254 CH933811 EDW06299.2 DS232047 EDS33064.1 CP012528 ALC48651.1 CH916371 EDV91735.1 KK853387 KDR07959.1 NNAY01000179 OXU30262.1 JXUM01016245 JXUM01016246 JXUM01016247 JXUM01016248 JXUM01016249 KQ560453 KXJ82341.1 KQ982430 KYQ56572.1 CH379063 EAL32401.3 JRES01000987 KNC26404.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000037510
UP000007151
UP000030742
+ More
UP000007266 UP000078541 UP000000311 UP000092462 UP000183832 UP000036403 UP000091820 UP000007798 UP000008711 UP000005205 UP000092443 UP000092460 UP000002282 UP000078542 UP000192221 UP000007801 UP000000803 UP000078492 UP000001292 UP000245037 UP000095301 UP000008792 UP000092444 UP000009192 UP000002320 UP000092553 UP000092445 UP000001070 UP000078200 UP000027135 UP000215335 UP000069940 UP000249989 UP000095300 UP000075809 UP000001819 UP000037069
UP000007266 UP000078541 UP000000311 UP000092462 UP000183832 UP000036403 UP000091820 UP000007798 UP000008711 UP000005205 UP000092443 UP000092460 UP000002282 UP000078542 UP000192221 UP000007801 UP000000803 UP000078492 UP000001292 UP000245037 UP000095301 UP000008792 UP000092444 UP000009192 UP000002320 UP000092553 UP000092445 UP000001070 UP000078200 UP000027135 UP000215335 UP000069940 UP000249989 UP000095300 UP000075809 UP000001819 UP000037069
Pfam
PF16061 DUF4803
Interpro
IPR032062
DUF4803
ProteinModelPortal
H9JXK3
A0A2H1VGY2
A0A194Q182
A0A2W1BM01
A0A2W1BMQ5
A0A0N1IPA6
+ More
A0A0L7KTD6 S4PWW1 A0A212EZ97 A0A1E1WRF0 A0A1E1WM67 U4U0Y7 A0A336MDK9 D6WVE3 A0A336MGF5 A0A195FMC4 A0A0A1WEF3 A0A1B6ENR6 E1ZZB9 A0A1B0D3K5 A0A1B6D7N1 A0A1B6C4N6 W8BRC6 A0A1Y1NB04 A0A1J1I244 A0A0J7NZ76 A0A1A9WCI2 A0A0Q9X3L3 B3NXZ9 A0A158NF43 A0A0K8V3H6 A0A034WPQ5 A0A1A9YI70 A0A1B0B2D2 B4Q0P7 A0A1W6EW95 A0A195C615 A0A1B6IJS5 A0A1W4VV31 A0A0K8U3Z3 B3MRI5 Q9W3M4 A0A195E952 B4IL60 A0A2P8YCW1 T1PCL3 A0A1I8M2B6 B4M713 A0A1B0GEG6 B4L540 B0WQX7 A0A0M4ET40 A0A1B0ACS7 B4JLX4 A0A1A9UZP5 A0A067QHF3 A0A232FHW8 A0A1B0AHW0 A0A182GR10 A0A1I8PL78 A0A151X876 A0A1B0AHV6 Q29JA0 A0A0L0C4P0
A0A0L7KTD6 S4PWW1 A0A212EZ97 A0A1E1WRF0 A0A1E1WM67 U4U0Y7 A0A336MDK9 D6WVE3 A0A336MGF5 A0A195FMC4 A0A0A1WEF3 A0A1B6ENR6 E1ZZB9 A0A1B0D3K5 A0A1B6D7N1 A0A1B6C4N6 W8BRC6 A0A1Y1NB04 A0A1J1I244 A0A0J7NZ76 A0A1A9WCI2 A0A0Q9X3L3 B3NXZ9 A0A158NF43 A0A0K8V3H6 A0A034WPQ5 A0A1A9YI70 A0A1B0B2D2 B4Q0P7 A0A1W6EW95 A0A195C615 A0A1B6IJS5 A0A1W4VV31 A0A0K8U3Z3 B3MRI5 Q9W3M4 A0A195E952 B4IL60 A0A2P8YCW1 T1PCL3 A0A1I8M2B6 B4M713 A0A1B0GEG6 B4L540 B0WQX7 A0A0M4ET40 A0A1B0ACS7 B4JLX4 A0A1A9UZP5 A0A067QHF3 A0A232FHW8 A0A1B0AHW0 A0A182GR10 A0A1I8PL78 A0A151X876 A0A1B0AHV6 Q29JA0 A0A0L0C4P0
Ontologies
KEGG
GO
Topology
SignalP
Position: 1 - 21,
Likelihood: 0.901855
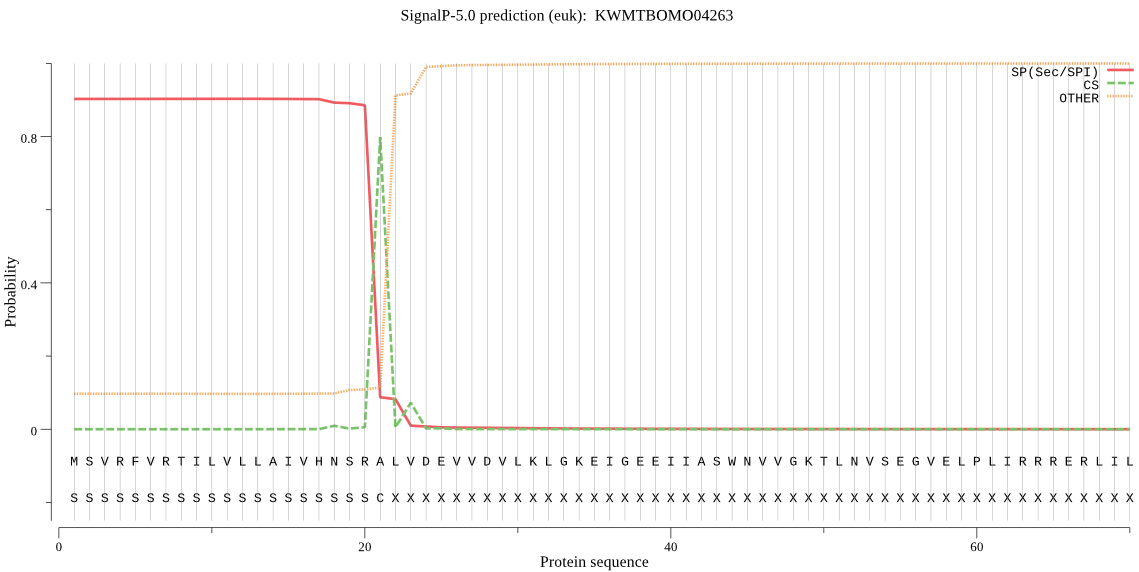
Length:
205
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0747600000000001
Exp number, first 60 AAs:
0.07409
Total prob of N-in:
0.08478
outside
1 - 205
Population Genetic Test Statistics
Pi
163.596122
Theta
192.824043
Tajima's D
-0.463641
CLR
0
CSRT
0.242837858107095
Interpretation
Uncertain
