Pre Gene Modal
BGIBMGA014236
Annotation
PREDICTED:_myosin-IA_[Papilio_xuthus]
Full name
Unconventional myosin ID
Alternative Name
Brush border myosin IA
Myosin 1D
Myosin-IA
Myosin 1D
Myosin-IA
Location in the cell
Mitochondrial Reliability : 2.032
Sequence
CDS
ATGCTCCTTTTATTCCAGGGCAGGGACACGTGCGTCCTTATATCGGGTGAATCAGGTGCAGGCAAAACTGAGGCATCTAAATTCATAATGAAGTACATCGCTGCGAACACAACACAAGTACACAGGGAATATATTGACAGAGTGAAAAACGTGTTGATACAGTCAAATACCATACTCGAAACATTTGGCAACGCAAAAACAAATCGCAACGATAATTCGTCCCGTTTCGGCAAATACATGGACATACATTTCGATTATAAGGGCGACCCAGTCGGCGGACACATCAGCAACTACCTCTTGGAGAAAAGTAGAGTGGTCAGCCTTCAACCGGGCGAGAGAAACTTTCATTCGTTTTATCAGCTCCTCAGTTCAAACAATCCGGTTTTGAAAAAACTGGGAATGTCAACGAACACGGCCTACAAGATCCTGGGCAGCGAAAGACCTACAGCGCACGATTCTAAACTTTATTCCCAAACGAATAGTGCGTTTAATGCTCTGGGCTTCTCTAATTCGGTCATCGAGGATATATGGAATATTGTCGCCGGTGTTATTTTATTGGGCGAGGTGACGTTCGAGGGCGGGGAGGGCGGGGAGGAGGGCGGCGCCGTGGCGGCGGGCGCGGTGGCGGGCGCGGTGCGTGCGCTGGGCGTGGCGGAGGCGGGGCTGCGCGCCGTGCTGGGGCGCCGCGTGCTGGCCGCCGGGCACGACCTGGTCACCAAGCAGCACTCGCTCGTGGACGCGCACTACACCAGGCTGGCGCTGGCCAAGGCGGCCTACGACAGACTCTTCACCTGGATCGTGCAACAGATAAACAAAGCCATCGACGTCCCGGCCGGCACGTACAAGAACACCCTCATCGGCGTGCTCGACATCTACGGGTTCGAGATCTTCGAGACCAACAGCTTCGAGCAGTTCTGCATCAACTACTGCAACGAGAAGTTGCAGCAGCTCTTCATCGAGCTGGTGCTGAAACAAGAGCAAGAGGAGTACGCGCGCGAGGGCATACAGTGGACGCCGGTGCAGTACTTCAACAACAGGATCATCTGCGAGCTCATCGACGCGCCCCATCACGGCGTCATCGCCATCATGGACGAGGCCTGCCTCAACCCCACACAGATCACGGACCAGCAGCTGCTGGAGGCGATGGACAAGCGGCTGACCGGCCACCGCCACTACACGTCTCGGCAGCTGGCGCCGACTGACAAGAAACTCAAGCACGGAGTCGACTTCAGGATAACGCACTACGCCGGCGACGTGACGTACGCCATCACCGGCTTCATGGACAAGAACAAGGACTCGCTGTGGCAGGACCTCAAGAGGCTGCTGCACTCCTCCAGCAACGCCTCGCTGGCCGCCATGTGGCCCGAGGGCGCCGCCGACATACAGAAGACCTCCCGGCGCCCCCCCTCCGCCGCGTCGCTGTTCCGCTCGTCGCTGGCGGCGCTGGTGGCGGGGCTGCACTCCAAGGAGCCCTTCTACGTGCGCTGCCTCAAGCCCAACCCCCTGCAGGCGCCCTCCACCTGGGACCCCCTGCTGGTGCAGCACCAGACGGCGTACCTGGGGCTGGTGGAGAACGTGCGCGTGCGACGGGCGGGGTTCGCGTGCCGCGTGCGCTACGACCGCTTCGTGCAGCGCTACAAGATGTTGTCGCAGTACACGTGGCCCAACTACCGCGGACACTCGCCCCGGGACGCCGCCGCCGTGCTGCTGCGGGACCTGCGCCTGGACGACGTGCACTACGGACACACCAAGCTCTTCATCAGGAGTGCTAAAACGCTGCAAAGCCTAGAGGCGGCCCGCGCAGAACTGATTCCTTCCATCGTGGTGCTGCTGCAGAAGTTGTGGCGAGGAGCGATCGCCAGGAGGAGATACAAGAAGATGAAGGCCGCACTAACCATATTCAACGCTTGGAAACGCTACCGCTACCGGCGCTACATCTCGGAGCTGCAGACGGAGCTGTCCCGGCACCGCGGCGTCATCCGGCGCTGGCCCCCCGCGCCGCGCGGCCTGGACCCCGCGCTGCTGGTGGCGGCGTACCGGCGGTGGCGCGCCTACCTCGTGCTGCGCGCCGTGCCGCGGGAGCGCTGGGCCGAGCTGCGCCTCAAGGTGTGCTGCGGCGCGGCGCTGCGGGGGCGGCGGCGGCACTGGGGCGCGGCCCGGCCGTGGCGCGGGGACTACCTGGCCGCGCCCGACTACAACGAGAAGGCGGGAGCGTACCGGGCGCAGGTCGCGAGCATGCAAAAGGCGGGTCAGATAGACAAGCCGCTGTTCTCGTGCCGCATACACAAGTTCAACCGCTACAACAAGTCCTCCGAGAGATGTCTGCTCGTCACTAGCAACGCGATATATAAATTGGATTCGAACAGTTTCAAGCCACTGAAGAAACCTACTTATATTAAAGACGTGGGGGCCGTGCGCGTGATGAGCTCGGACGTGCAGCTGGTGGTGCTGTGCGTGCCGTCCGCCAAGAACGACCTGGTGGCCGCGCTCGAAGCCCCCGGGGACGACCTGGTCGGCGAGCTGCTGGGCGTGCTGGCCAGCAGATACTGCATGTTGTCTGGCGGCGAGCTGTGCGTGGAGGTGGAGAGCGGCGCCAGCACGCGCTGCACGCTGGGGGGCCGCAGCCGCGCGCTGCAGCTGCCCGCGCACGCCGACCCGCGCGCGCCCGCCTTCACGCACGCGCACAACGTCATCACCTACCACCCGCCGTCCGCGCGCGCCTGA
Protein
MLLLFQGRDTCVLISGESGAGKTEASKFIMKYIAANTTQVHREYIDRVKNVLIQSNTILETFGNAKTNRNDNSSRFGKYMDIHFDYKGDPVGGHISNYLLEKSRVVSLQPGERNFHSFYQLLSSNNPVLKKLGMSTNTAYKILGSERPTAHDSKLYSQTNSAFNALGFSNSVIEDIWNIVAGVILLGEVTFEGGEGGEEGGAVAAGAVAGAVRALGVAEAGLRAVLGRRVLAAGHDLVTKQHSLVDAHYTRLALAKAAYDRLFTWIVQQINKAIDVPAGTYKNTLIGVLDIYGFEIFETNSFEQFCINYCNEKLQQLFIELVLKQEQEEYAREGIQWTPVQYFNNRIICELIDAPHHGVIAIMDEACLNPTQITDQQLLEAMDKRLTGHRHYTSRQLAPTDKKLKHGVDFRITHYAGDVTYAITGFMDKNKDSLWQDLKRLLHSSSNASLAAMWPEGAADIQKTSRRPPSAASLFRSSLAALVAGLHSKEPFYVRCLKPNPLQAPSTWDPLLVQHQTAYLGLVENVRVRRAGFACRVRYDRFVQRYKMLSQYTWPNYRGHSPRDAAAVLLRDLRLDDVHYGHTKLFIRSAKTLQSLEAARAELIPSIVVLLQKLWRGAIARRRYKKMKAALTIFNAWKRYRYRRYISELQTELSRHRGVIRRWPPAPRGLDPALLVAAYRRWRAYLVLRAVPRERWAELRLKVCCGAALRGRRRHWGAARPWRGDYLAAPDYNEKAGAYRAQVASMQKAGQIDKPLFSCRIHKFNRYNKSSERCLLVTSNAIYKLDSNSFKPLKKPTYIKDVGAVRVMSSDVQLVVLCVPSAKNDLVAALEAPGDDLVGELLGVLASRYCMLSGGELCVEVESGASTRCTLGGRSRALQLPAHADPRAPAFTHAHNVITYHPPSARA
Summary
Description
Unconventional myosin that functions as actin-based motor protein with ATPase activity (PubMed:30467170). Binds to membranes enriched in phosphatidylinositol 4-5-bisphosphate, and can glide along actin filaments when anchored to a lipid bilayer (PubMed:30467170). Generates left-right asymmetry at the level of single cells, organs and the whole body via its interaction with the actin cytoskeleton, both in the embryo and the adult (PubMed:16598258, PubMed:16598259, PubMed:18521948, PubMed:26073018, PubMed:25659376, PubMed:30467170). Normal left-right asymmetry of the larval midgut and hindgut requires expression in the embryonic hindgut epithelium during a critical time period, 10 to 12.75 hours after egg laying (PubMed:18521948). This period corresponds to a late stage of germband retraction, and precedes left-right asymmetric morphogenesis (PubMed:18521948). Expression in segment H1 of the imaginal ring is required at 0 to 24 hours after pupation for changes of cell shape and orientation in the H2 segment, which then gives rise to normal, dextral looping of the adult hindgut (PubMed:16598258, PubMed:26073018). Required during a critical period, 126-132 hours after egg laying, for normal, dextral rotation of the adult male genitalia (PubMed:16598258, PubMed:16598259, PubMed:22491943, PubMed:26073018, PubMed:25659376). Has a double role by promoting dextral rotation in the posterior compartment of segment A8 of the male genital disk, and in repressing sinistral looping in the anterior compartment (PubMed:16598259).
Subunit
Binds to F-actin (PubMed:7589814). Interacts with arm (PubMed:16598259, PubMed:22491943). Interacts with shg (PubMed:22491943). Interacts with ds (via intracellular region) (PubMed:26073018).
Miscellaneous
Overexpression throughout the embryo reverses the normal left-right asymmetry of the embryonic gut, giving rise to a gut that is a mirror-image of the wild-type (PubMed:16598258). Ectopic expression in the larval epidermis causes dextral twisting of the entire larval body. Ectopic expression in tracheal precursor cells causes dextral spiraling of tracheal branches, giving rise to a spiraling ribbon shape instead of the normal smooth tube. Ectopic expression in epithelial cells causes increased elongation and a clear shift of the membrane orientation toward one side, so that the membrane is no longer perpendicular to the anterior-posterior axis (PubMed:30467170).
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Keywords
Actin-binding
ATP-binding
Calmodulin-binding
Cell junction
Cell membrane
Cell projection
Complete proteome
Cytoplasm
Cytoskeleton
Developmental protein
Lipid-binding
Membrane
Motor protein
Myosin
Nucleotide-binding
Reference proteome
Repeat
Feature
chain Unconventional myosin ID
Uniprot
A0A3S2NAI9
A0A2W1BFQ0
A0A2H1VGM8
A0A2P8Y3B6
A0A2J7PHM3
A0A212FGN4
+ More
A0A067QRJ8 A0A1Y1LB74 E0VG03 A0A348G643 A0A146M4P3 A0A1B6DEF2 A0A0A9YGP8 E9IBH9 A0A069DWZ1 D6WVP8 A0A151I573 A0A0C9QM31 A0A158NI22 A0A023F3X7 T1PIF4 A0A195EFU6 A0A224XIN5 U5EJT2 E2APC4 A0A195CLC6 A0A151WJT1 A0A3L8D8Q5 A0A2A3E5N1 A0A195F791 A0A232FGS1 A0A0A1WYK4 K7IQ81 A0A088A019 A0A0L0BSF7 A0A3B0K6Y6 A0A1I8PHP1 A0A034WM34 A0A0M9A9S4 E2C8J5 B4LTG9 B4JDW9 B5DIK4 B4G9R7 A0A0K8VTB2 B4KG68 A0A0K8WA16 Q16LJ3 A0A182ILA2 B3MJJ7 Q23978 A0A0C9PT95 B3N5E4 A0A0P6G8S4 A0A182K1R8 B4N2R4 A0A0P5CZH5 B4P0Q4 A0A0P5CZ12 A0A0L7R7D4 A0A182KJL8 A0A0P5A8U7 B0WED3 A0A1S4H370 A0A182HQU7 N6TQE2 A0A182XLG1 A0A0M4EQZ2 A0A182U3T9 A0A182FMY8 A0A1J1I120 B4Q9C1 W8C5Z0 A0A1B0A0M6 A0A182V1H0 A0A0P5TLN7 A0A182MQJ1 A0A0P5ZJ96 A0A1B0G658 A0A1A9Y4Q5 W5JAZ0 A0A182PCH4 A0A182WA60 A0A182YIL6 A0A182QJU2 A0A0P6HC55 A0A2M4A447 A0A2M4A4C6 A0A1W4UFK5 A0A182R7R0 A0A0P5MRR9 A0A131XZ66 A0A147BHG0 A0A1E1XPT8 A0A131Z2L1 A0A224Z3N2 A0A0N8BRF6 A0A1A9UX91 A0A131XP45
A0A067QRJ8 A0A1Y1LB74 E0VG03 A0A348G643 A0A146M4P3 A0A1B6DEF2 A0A0A9YGP8 E9IBH9 A0A069DWZ1 D6WVP8 A0A151I573 A0A0C9QM31 A0A158NI22 A0A023F3X7 T1PIF4 A0A195EFU6 A0A224XIN5 U5EJT2 E2APC4 A0A195CLC6 A0A151WJT1 A0A3L8D8Q5 A0A2A3E5N1 A0A195F791 A0A232FGS1 A0A0A1WYK4 K7IQ81 A0A088A019 A0A0L0BSF7 A0A3B0K6Y6 A0A1I8PHP1 A0A034WM34 A0A0M9A9S4 E2C8J5 B4LTG9 B4JDW9 B5DIK4 B4G9R7 A0A0K8VTB2 B4KG68 A0A0K8WA16 Q16LJ3 A0A182ILA2 B3MJJ7 Q23978 A0A0C9PT95 B3N5E4 A0A0P6G8S4 A0A182K1R8 B4N2R4 A0A0P5CZH5 B4P0Q4 A0A0P5CZ12 A0A0L7R7D4 A0A182KJL8 A0A0P5A8U7 B0WED3 A0A1S4H370 A0A182HQU7 N6TQE2 A0A182XLG1 A0A0M4EQZ2 A0A182U3T9 A0A182FMY8 A0A1J1I120 B4Q9C1 W8C5Z0 A0A1B0A0M6 A0A182V1H0 A0A0P5TLN7 A0A182MQJ1 A0A0P5ZJ96 A0A1B0G658 A0A1A9Y4Q5 W5JAZ0 A0A182PCH4 A0A182WA60 A0A182YIL6 A0A182QJU2 A0A0P6HC55 A0A2M4A447 A0A2M4A4C6 A0A1W4UFK5 A0A182R7R0 A0A0P5MRR9 A0A131XZ66 A0A147BHG0 A0A1E1XPT8 A0A131Z2L1 A0A224Z3N2 A0A0N8BRF6 A0A1A9UX91 A0A131XP45
Pubmed
28756777
29403074
22118469
24845553
28004739
20566863
+ More
26823975 25401762 21282665 26334808 18362917 19820115 21347285 25474469 25315136 20798317 30249741 28648823 25830018 20075255 26108605 25348373 17994087 15632085 17510324 8201616 10731132 12537572 12537569 7589814 16598258 16598259 18521948 22491943 26073018 25659376 30467170 18057021 17550304 20966253 12364791 23537049 22936249 24495485 20920257 23761445 25244985 29652888 29209593 26830274 28797301 28049606
26823975 25401762 21282665 26334808 18362917 19820115 21347285 25474469 25315136 20798317 30249741 28648823 25830018 20075255 26108605 25348373 17994087 15632085 17510324 8201616 10731132 12537572 12537569 7589814 16598258 16598259 18521948 22491943 26073018 25659376 30467170 18057021 17550304 20966253 12364791 23537049 22936249 24495485 20920257 23761445 25244985 29652888 29209593 26830274 28797301 28049606
EMBL
RSAL01000147
RVE45880.1
KZ150080
PZC73892.1
ODYU01002476
SOQ40010.1
+ More
PYGN01000977 PSN38767.1 NEVH01025135 PNF15832.1 AGBW02008642 OWR52887.1 KK853042 KDR12139.1 GEZM01060826 JAV70873.1 DS235129 EEB12309.1 FX985583 BBF97916.1 GDHC01016891 GDHC01004627 JAQ01738.1 JAQ14002.1 GEDC01018061 GEDC01013323 GEDC01009007 GEDC01000987 JAS19237.1 JAS23975.1 JAS28291.1 JAS36311.1 GBHO01036065 GBHO01012793 GBHO01012762 JAG07539.1 JAG30811.1 JAG30842.1 GL762111 EFZ22181.1 GBGD01000296 JAC88593.1 KQ971358 EFA08265.1 KQ976456 KYM84877.1 GBYB01004594 JAG74361.1 ADTU01016187 ADTU01016188 GBBI01002562 JAC16150.1 KA647910 AFP62539.1 KQ978957 KYN27145.1 GFTR01008433 JAW07993.1 GANO01002110 JAB57761.1 GL441504 EFN64723.1 KQ977622 KYN01267.1 KQ983039 KYQ48060.1 QOIP01000012 RLU16318.1 KZ288379 PBC26592.1 KQ981763 KYN36052.1 NNAY01000281 OXU29527.1 GBXI01010143 JAD04149.1 JRES01001427 KNC22977.1 OUUW01000004 SPP79258.1 GAKP01003527 GAKP01003526 GAKP01003525 JAC55427.1 KQ435719 KOX78853.1 GL453666 EFN75725.1 CH940649 EDW63939.1 CH916368 EDW03489.1 CH379060 EDY70146.1 CH479180 EDW29097.1 GDHF01024750 GDHF01010173 JAI27564.1 JAI42141.1 CH933807 EDW11055.1 GDHF01004281 JAI48033.1 CH477904 EAT35186.1 CH902620 EDV32365.1 U07595 AE014134 AY075567 GBYB01004593 JAG74360.1 CH954177 EDV59023.1 KQS70598.1 GDIQ01036955 JAN57782.1 CH963977 EDW78653.1 GDIP01179415 JAJ43987.1 CM000157 EDW87949.1 KRJ97505.1 GDIP01252461 GDIP01167675 LRGB01000930 JAJ55727.1 KZS14906.1 KQ414643 KOC66671.1 GDIP01214862 JAJ08540.1 DS231907 EDS45493.1 AAAB01008980 APCN01001600 APGK01058605 APGK01058606 KB741291 ENN70521.1 CP012523 ALC39232.1 CVRI01000038 CRK94047.1 CM000361 CM002910 EDX04563.1 KMY89567.1 GAMC01004616 GAMC01004615 GAMC01004614 GAMC01004613 JAC01942.1 GDIP01129930 JAL73784.1 AXCM01000205 GDIP01044289 JAM59426.1 CCAG010007078 ADMH02002025 ETN59929.1 AXCN02001858 GDIQ01021319 JAN73418.1 GGFK01002179 MBW35500.1 GGFK01002177 MBW35498.1 GDIQ01152212 JAK99513.1 GEFM01004248 JAP71548.1 GEGO01005174 JAR90230.1 GFAA01002159 JAU01276.1 GEDV01003369 JAP85188.1 GFPF01010027 MAA21173.1 GDIQ01152211 JAK99514.1 GEFH01000691 JAP67890.1
PYGN01000977 PSN38767.1 NEVH01025135 PNF15832.1 AGBW02008642 OWR52887.1 KK853042 KDR12139.1 GEZM01060826 JAV70873.1 DS235129 EEB12309.1 FX985583 BBF97916.1 GDHC01016891 GDHC01004627 JAQ01738.1 JAQ14002.1 GEDC01018061 GEDC01013323 GEDC01009007 GEDC01000987 JAS19237.1 JAS23975.1 JAS28291.1 JAS36311.1 GBHO01036065 GBHO01012793 GBHO01012762 JAG07539.1 JAG30811.1 JAG30842.1 GL762111 EFZ22181.1 GBGD01000296 JAC88593.1 KQ971358 EFA08265.1 KQ976456 KYM84877.1 GBYB01004594 JAG74361.1 ADTU01016187 ADTU01016188 GBBI01002562 JAC16150.1 KA647910 AFP62539.1 KQ978957 KYN27145.1 GFTR01008433 JAW07993.1 GANO01002110 JAB57761.1 GL441504 EFN64723.1 KQ977622 KYN01267.1 KQ983039 KYQ48060.1 QOIP01000012 RLU16318.1 KZ288379 PBC26592.1 KQ981763 KYN36052.1 NNAY01000281 OXU29527.1 GBXI01010143 JAD04149.1 JRES01001427 KNC22977.1 OUUW01000004 SPP79258.1 GAKP01003527 GAKP01003526 GAKP01003525 JAC55427.1 KQ435719 KOX78853.1 GL453666 EFN75725.1 CH940649 EDW63939.1 CH916368 EDW03489.1 CH379060 EDY70146.1 CH479180 EDW29097.1 GDHF01024750 GDHF01010173 JAI27564.1 JAI42141.1 CH933807 EDW11055.1 GDHF01004281 JAI48033.1 CH477904 EAT35186.1 CH902620 EDV32365.1 U07595 AE014134 AY075567 GBYB01004593 JAG74360.1 CH954177 EDV59023.1 KQS70598.1 GDIQ01036955 JAN57782.1 CH963977 EDW78653.1 GDIP01179415 JAJ43987.1 CM000157 EDW87949.1 KRJ97505.1 GDIP01252461 GDIP01167675 LRGB01000930 JAJ55727.1 KZS14906.1 KQ414643 KOC66671.1 GDIP01214862 JAJ08540.1 DS231907 EDS45493.1 AAAB01008980 APCN01001600 APGK01058605 APGK01058606 KB741291 ENN70521.1 CP012523 ALC39232.1 CVRI01000038 CRK94047.1 CM000361 CM002910 EDX04563.1 KMY89567.1 GAMC01004616 GAMC01004615 GAMC01004614 GAMC01004613 JAC01942.1 GDIP01129930 JAL73784.1 AXCM01000205 GDIP01044289 JAM59426.1 CCAG010007078 ADMH02002025 ETN59929.1 AXCN02001858 GDIQ01021319 JAN73418.1 GGFK01002179 MBW35500.1 GGFK01002177 MBW35498.1 GDIQ01152212 JAK99513.1 GEFM01004248 JAP71548.1 GEGO01005174 JAR90230.1 GFAA01002159 JAU01276.1 GEDV01003369 JAP85188.1 GFPF01010027 MAA21173.1 GDIQ01152211 JAK99514.1 GEFH01000691 JAP67890.1
Proteomes
UP000283053
UP000245037
UP000235965
UP000007151
UP000027135
UP000009046
+ More
UP000007266 UP000078540 UP000005205 UP000095301 UP000078492 UP000000311 UP000078542 UP000075809 UP000279307 UP000242457 UP000078541 UP000215335 UP000002358 UP000005203 UP000037069 UP000268350 UP000095300 UP000053105 UP000008237 UP000008792 UP000001070 UP000001819 UP000008744 UP000009192 UP000008820 UP000075880 UP000007801 UP000000803 UP000008711 UP000075881 UP000007798 UP000002282 UP000076858 UP000053825 UP000075882 UP000002320 UP000075840 UP000019118 UP000076407 UP000092553 UP000075902 UP000069272 UP000183832 UP000000304 UP000092445 UP000075903 UP000075883 UP000092444 UP000092443 UP000000673 UP000075885 UP000075920 UP000076408 UP000075886 UP000192221 UP000075900 UP000078200
UP000007266 UP000078540 UP000005205 UP000095301 UP000078492 UP000000311 UP000078542 UP000075809 UP000279307 UP000242457 UP000078541 UP000215335 UP000002358 UP000005203 UP000037069 UP000268350 UP000095300 UP000053105 UP000008237 UP000008792 UP000001070 UP000001819 UP000008744 UP000009192 UP000008820 UP000075880 UP000007801 UP000000803 UP000008711 UP000075881 UP000007798 UP000002282 UP000076858 UP000053825 UP000075882 UP000002320 UP000075840 UP000019118 UP000076407 UP000092553 UP000075902 UP000069272 UP000183832 UP000000304 UP000092445 UP000075903 UP000075883 UP000092444 UP000092443 UP000000673 UP000075885 UP000075920 UP000076408 UP000075886 UP000192221 UP000075900 UP000078200
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
A0A3S2NAI9
A0A2W1BFQ0
A0A2H1VGM8
A0A2P8Y3B6
A0A2J7PHM3
A0A212FGN4
+ More
A0A067QRJ8 A0A1Y1LB74 E0VG03 A0A348G643 A0A146M4P3 A0A1B6DEF2 A0A0A9YGP8 E9IBH9 A0A069DWZ1 D6WVP8 A0A151I573 A0A0C9QM31 A0A158NI22 A0A023F3X7 T1PIF4 A0A195EFU6 A0A224XIN5 U5EJT2 E2APC4 A0A195CLC6 A0A151WJT1 A0A3L8D8Q5 A0A2A3E5N1 A0A195F791 A0A232FGS1 A0A0A1WYK4 K7IQ81 A0A088A019 A0A0L0BSF7 A0A3B0K6Y6 A0A1I8PHP1 A0A034WM34 A0A0M9A9S4 E2C8J5 B4LTG9 B4JDW9 B5DIK4 B4G9R7 A0A0K8VTB2 B4KG68 A0A0K8WA16 Q16LJ3 A0A182ILA2 B3MJJ7 Q23978 A0A0C9PT95 B3N5E4 A0A0P6G8S4 A0A182K1R8 B4N2R4 A0A0P5CZH5 B4P0Q4 A0A0P5CZ12 A0A0L7R7D4 A0A182KJL8 A0A0P5A8U7 B0WED3 A0A1S4H370 A0A182HQU7 N6TQE2 A0A182XLG1 A0A0M4EQZ2 A0A182U3T9 A0A182FMY8 A0A1J1I120 B4Q9C1 W8C5Z0 A0A1B0A0M6 A0A182V1H0 A0A0P5TLN7 A0A182MQJ1 A0A0P5ZJ96 A0A1B0G658 A0A1A9Y4Q5 W5JAZ0 A0A182PCH4 A0A182WA60 A0A182YIL6 A0A182QJU2 A0A0P6HC55 A0A2M4A447 A0A2M4A4C6 A0A1W4UFK5 A0A182R7R0 A0A0P5MRR9 A0A131XZ66 A0A147BHG0 A0A1E1XPT8 A0A131Z2L1 A0A224Z3N2 A0A0N8BRF6 A0A1A9UX91 A0A131XP45
A0A067QRJ8 A0A1Y1LB74 E0VG03 A0A348G643 A0A146M4P3 A0A1B6DEF2 A0A0A9YGP8 E9IBH9 A0A069DWZ1 D6WVP8 A0A151I573 A0A0C9QM31 A0A158NI22 A0A023F3X7 T1PIF4 A0A195EFU6 A0A224XIN5 U5EJT2 E2APC4 A0A195CLC6 A0A151WJT1 A0A3L8D8Q5 A0A2A3E5N1 A0A195F791 A0A232FGS1 A0A0A1WYK4 K7IQ81 A0A088A019 A0A0L0BSF7 A0A3B0K6Y6 A0A1I8PHP1 A0A034WM34 A0A0M9A9S4 E2C8J5 B4LTG9 B4JDW9 B5DIK4 B4G9R7 A0A0K8VTB2 B4KG68 A0A0K8WA16 Q16LJ3 A0A182ILA2 B3MJJ7 Q23978 A0A0C9PT95 B3N5E4 A0A0P6G8S4 A0A182K1R8 B4N2R4 A0A0P5CZH5 B4P0Q4 A0A0P5CZ12 A0A0L7R7D4 A0A182KJL8 A0A0P5A8U7 B0WED3 A0A1S4H370 A0A182HQU7 N6TQE2 A0A182XLG1 A0A0M4EQZ2 A0A182U3T9 A0A182FMY8 A0A1J1I120 B4Q9C1 W8C5Z0 A0A1B0A0M6 A0A182V1H0 A0A0P5TLN7 A0A182MQJ1 A0A0P5ZJ96 A0A1B0G658 A0A1A9Y4Q5 W5JAZ0 A0A182PCH4 A0A182WA60 A0A182YIL6 A0A182QJU2 A0A0P6HC55 A0A2M4A447 A0A2M4A4C6 A0A1W4UFK5 A0A182R7R0 A0A0P5MRR9 A0A131XZ66 A0A147BHG0 A0A1E1XPT8 A0A131Z2L1 A0A224Z3N2 A0A0N8BRF6 A0A1A9UX91 A0A131XP45
PDB
5V7X
E-value=2.31603e-134,
Score=1230
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Detected throughout the cytoplasm. Detected primarily in the basolateral cytoplasmic region of immature enterocytes. Detected also in the apical cytoplasmic region in midgut enterocytes and follicle cells (PubMed:7589814). Colocalizes with the actin cytoskeleton (PubMed:7589814, PubMed:16598258, PubMed:16598259, PubMed:25659376). Colocalizes with arm at adherens junctions (PubMed:16598259). Colocalizes with ds at cell junctions (PubMed:26073018). With evidence from 4 publications.
Cell cortex Detected throughout the cytoplasm. Detected primarily in the basolateral cytoplasmic region of immature enterocytes. Detected also in the apical cytoplasmic region in midgut enterocytes and follicle cells (PubMed:7589814). Colocalizes with the actin cytoskeleton (PubMed:7589814, PubMed:16598258, PubMed:16598259, PubMed:25659376). Colocalizes with arm at adherens junctions (PubMed:16598259). Colocalizes with ds at cell junctions (PubMed:26073018). With evidence from 4 publications.
Cytoskeleton Detected throughout the cytoplasm. Detected primarily in the basolateral cytoplasmic region of immature enterocytes. Detected also in the apical cytoplasmic region in midgut enterocytes and follicle cells (PubMed:7589814). Colocalizes with the actin cytoskeleton (PubMed:7589814, PubMed:16598258, PubMed:16598259, PubMed:25659376). Colocalizes with arm at adherens junctions (PubMed:16598259). Colocalizes with ds at cell junctions (PubMed:26073018). With evidence from 4 publications.
Cell membrane Detected throughout the cytoplasm. Detected primarily in the basolateral cytoplasmic region of immature enterocytes. Detected also in the apical cytoplasmic region in midgut enterocytes and follicle cells (PubMed:7589814). Colocalizes with the actin cytoskeleton (PubMed:7589814, PubMed:16598258, PubMed:16598259, PubMed:25659376). Colocalizes with arm at adherens junctions (PubMed:16598259). Colocalizes with ds at cell junctions (PubMed:26073018). With evidence from 4 publications.
Cell junction Detected throughout the cytoplasm. Detected primarily in the basolateral cytoplasmic region of immature enterocytes. Detected also in the apical cytoplasmic region in midgut enterocytes and follicle cells (PubMed:7589814). Colocalizes with the actin cytoskeleton (PubMed:7589814, PubMed:16598258, PubMed:16598259, PubMed:25659376). Colocalizes with arm at adherens junctions (PubMed:16598259). Colocalizes with ds at cell junctions (PubMed:26073018). With evidence from 4 publications.
Adherens junction Detected throughout the cytoplasm. Detected primarily in the basolateral cytoplasmic region of immature enterocytes. Detected also in the apical cytoplasmic region in midgut enterocytes and follicle cells (PubMed:7589814). Colocalizes with the actin cytoskeleton (PubMed:7589814, PubMed:16598258, PubMed:16598259, PubMed:25659376). Colocalizes with arm at adherens junctions (PubMed:16598259). Colocalizes with ds at cell junctions (PubMed:26073018). With evidence from 4 publications.
Cell projection Detected throughout the cytoplasm. Detected primarily in the basolateral cytoplasmic region of immature enterocytes. Detected also in the apical cytoplasmic region in midgut enterocytes and follicle cells (PubMed:7589814). Colocalizes with the actin cytoskeleton (PubMed:7589814, PubMed:16598258, PubMed:16598259, PubMed:25659376). Colocalizes with arm at adherens junctions (PubMed:16598259). Colocalizes with ds at cell junctions (PubMed:26073018). With evidence from 4 publications.
Cell cortex Detected throughout the cytoplasm. Detected primarily in the basolateral cytoplasmic region of immature enterocytes. Detected also in the apical cytoplasmic region in midgut enterocytes and follicle cells (PubMed:7589814). Colocalizes with the actin cytoskeleton (PubMed:7589814, PubMed:16598258, PubMed:16598259, PubMed:25659376). Colocalizes with arm at adherens junctions (PubMed:16598259). Colocalizes with ds at cell junctions (PubMed:26073018). With evidence from 4 publications.
Cytoskeleton Detected throughout the cytoplasm. Detected primarily in the basolateral cytoplasmic region of immature enterocytes. Detected also in the apical cytoplasmic region in midgut enterocytes and follicle cells (PubMed:7589814). Colocalizes with the actin cytoskeleton (PubMed:7589814, PubMed:16598258, PubMed:16598259, PubMed:25659376). Colocalizes with arm at adherens junctions (PubMed:16598259). Colocalizes with ds at cell junctions (PubMed:26073018). With evidence from 4 publications.
Cell membrane Detected throughout the cytoplasm. Detected primarily in the basolateral cytoplasmic region of immature enterocytes. Detected also in the apical cytoplasmic region in midgut enterocytes and follicle cells (PubMed:7589814). Colocalizes with the actin cytoskeleton (PubMed:7589814, PubMed:16598258, PubMed:16598259, PubMed:25659376). Colocalizes with arm at adherens junctions (PubMed:16598259). Colocalizes with ds at cell junctions (PubMed:26073018). With evidence from 4 publications.
Cell junction Detected throughout the cytoplasm. Detected primarily in the basolateral cytoplasmic region of immature enterocytes. Detected also in the apical cytoplasmic region in midgut enterocytes and follicle cells (PubMed:7589814). Colocalizes with the actin cytoskeleton (PubMed:7589814, PubMed:16598258, PubMed:16598259, PubMed:25659376). Colocalizes with arm at adherens junctions (PubMed:16598259). Colocalizes with ds at cell junctions (PubMed:26073018). With evidence from 4 publications.
Adherens junction Detected throughout the cytoplasm. Detected primarily in the basolateral cytoplasmic region of immature enterocytes. Detected also in the apical cytoplasmic region in midgut enterocytes and follicle cells (PubMed:7589814). Colocalizes with the actin cytoskeleton (PubMed:7589814, PubMed:16598258, PubMed:16598259, PubMed:25659376). Colocalizes with arm at adherens junctions (PubMed:16598259). Colocalizes with ds at cell junctions (PubMed:26073018). With evidence from 4 publications.
Cell projection Detected throughout the cytoplasm. Detected primarily in the basolateral cytoplasmic region of immature enterocytes. Detected also in the apical cytoplasmic region in midgut enterocytes and follicle cells (PubMed:7589814). Colocalizes with the actin cytoskeleton (PubMed:7589814, PubMed:16598258, PubMed:16598259, PubMed:25659376). Colocalizes with arm at adherens junctions (PubMed:16598259). Colocalizes with ds at cell junctions (PubMed:26073018). With evidence from 4 publications.
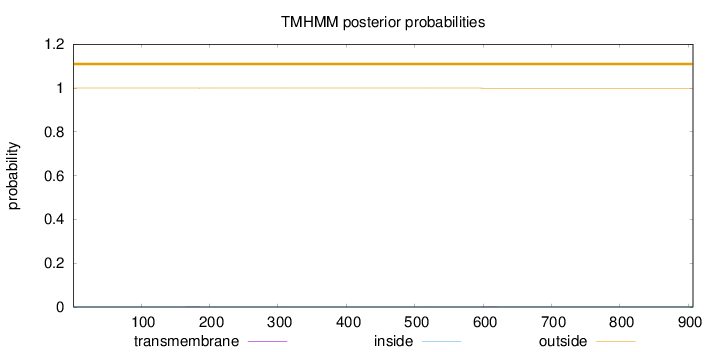
Length:
907
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02369
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00020
outside
1 - 907
Population Genetic Test Statistics
Pi
267.599419
Theta
191.415808
Tajima's D
1.129482
CLR
0.126555
CSRT
0.692265386730664
Interpretation
Uncertain
