Gene
KWMTBOMO04254
Pre Gene Modal
BGIBMGA014266
Annotation
PREDICTED:_cationic_amino_acid_transporter_2_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 4.955
Sequence
CDS
ATGGGCTGCGGTAAGATCTTCGCGACGCTACGTCGGTGCAAAAAAATTGATTATGACGACGACACCACCCAGTTGTCCAGATGTCTTGGACTATTCGACCTCACCTCTCTAGGAGTCGGGAGTACCCTCGGTCTAGGAGTGTACGTGCTGGCCGGCGCTGTCGCCAAGACTGTTGCTGGACCGGCGGTTGCCATCAGCTTTTTGGTGGCAGCCATTTCTTCTGTATTTGCTGGGTTATGTTACGCAGAATTCGCTTCAAGAGTCCCAAAAGCCGGTTCAGGCTATAACTACAGTTACGTGAGCGTCGGGGAGTTCATCGCGTTCACGATTGGTTGGAACTTGATCTTGGAGTACGCTATAGGAACAGCCAGCGTCGCCAAAGGCATGGCGGTCTACATTGACACCCTCTTCAATAACACCATGGCGAAAACCTTGACCGCGGCTACTCCGATCAATGTGTCCTTCTTGGCTGACTACCCTGATTTCTTTGCATTTGGTCTAGTTTTTTTGGTAACTATTCTGCTCGGTATTGGAGTTAACGAATCTGCGAAAATGAATAACGTATTCACGGCCCTCAATGTATTAACTGTCATAGTTGTTGTCGTCGCGGGCGCTGTCAAAAGTAACCCAGCTAACTGGAACATCAAACTAGCCGATATTCCACCCGAGTACGTGAGTCAAGCCGGGGAAGGAGGCTTTATGCCGTGGGGTGTGGCGGGCGTCATGGCGGGAGCGGCTAAATGCTTCTTCGGCTTCGTCGGCTTCGACTGCGTGGCCACCACGGGGGAAGAGGCCAAGAACCCGAAAAGAGACATACCTCTGTCAATAGTTCTGTCTCTGATCATAATCTTCTTGTCATATTTCTCCATCGCGACAGTGCTCACCATGATGCTGCCTTATTATATGCAGGACGCGGAGGCCCCATTCCCCTACGTATTCGAACAGGTTAACTTACCGGTGATCAAGTGGGTGGTGACAGTGGGTGCTATATTCGCTTTGTGCACGAGTCTGCTCGGCACCATGTTCCCACTCCCGCGGGTGCTGTACGCTATGGCGTGTGACGGGGTGCTGTTCCGACCACTAGCCGCCATTCACCCGAAGACCAAGACCCCATTGTTGGCCACTGCGATTAGTGGATTTCTTTCGGCAATAATGGCAGCCATGTTCAATCTGAAACAACTGATAGACATGGTGTCTATTGGGACTTTGCTAGCGTACACCATCGTGGCAACAAGCGTTCTCATACTTAGATACGAGGAGGAGGATGTATCTGTTGGCACGGAAAAAATATCGGCACGCGGGAGGGGAGTGCTAGCAGTATTAGGTCAGGGTTGCAACCTGCTCGGACTCAAGAACCCCACGCAACTCTCCTCCACCATCGCCAAGATTACCATCGCTGTCCTCTTCGTGGTGGCGCTGGCGACGTGCGTAGTATTGAGTGTGGACGGCGGAGCGGTGGCGGGAGCAATACTGGGGGCGATCCTGGTGGTATTACTGGTGGTGTTATACCGACAACCCACTAATGACGTATCCCATCTCTCATTCAAGGTGCCGCTAGTCCCACTTGTCCCATACCTGAGCGTCTGCATGAACGTGTATCTGATGGTCCAGTTGGACTACCAGACGTGGGTTCGGTTTTTCATATGGCTTATAATCGGTTACCTAATTTACTTCTTATATGGAATCCGTAACAGTTCACTCAATTCTTCACAGCCCGCCGGTAAAGTCGATGACAAGGATAAATCACAAGTCGTAACAAAATTTTGA
Protein
MGCGKIFATLRRCKKIDYDDDTTQLSRCLGLFDLTSLGVGSTLGLGVYVLAGAVAKTVAGPAVAISFLVAAISSVFAGLCYAEFASRVPKAGSGYNYSYVSVGEFIAFTIGWNLILEYAIGTASVAKGMAVYIDTLFNNTMAKTLTAATPINVSFLADYPDFFAFGLVFLVTILLGIGVNESAKMNNVFTALNVLTVIVVVVAGAVKSNPANWNIKLADIPPEYVSQAGEGGFMPWGVAGVMAGAAKCFFGFVGFDCVATTGEEAKNPKRDIPLSIVLSLIIIFLSYFSIATVLTMMLPYYMQDAEAPFPYVFEQVNLPVIKWVVTVGAIFALCTSLLGTMFPLPRVLYAMACDGVLFRPLAAIHPKTKTPLLATAISGFLSAIMAAMFNLKQLIDMVSIGTLLAYTIVATSVLILRYEEEDVSVGTEKISARGRGVLAVLGQGCNLLGLKNPTQLSSTIAKITIAVLFVVALATCVVLSVDGGAVAGAILGAILVVLLVVLYRQPTNDVSHLSFKVPLVPLVPYLSVCMNVYLMVQLDYQTWVRFFIWLIIGYLIYFLYGIRNSSLNSSQPAGKVDDKDKSQVVTKF
Summary
Uniprot
H9JXK0
H9JXJ9
A0A3S2TH46
A0A2H1VXW1
A0A194QMV8
A0A194Q172
+ More
A0A212F0U1 A0A2A4IUH9 A0A026X573 A0A310STV1 A0A2W1BFK7 A0A2A4IX97 A0A2A3ERP8 A0A195ET83 A0A151I7E1 K7IVG0 E9IB29 A0A0M9ACZ5 A0A158NVX0 A0A151WSG4 F4X4X5 A0A067RRF9 A0A336LKG1 A0A0L7QXA2 A0A151HYS8 B4J3S2 A0A1I8MEN9 A0A1J1HMV0 A0A1Y1MQL8 A0A1Y1MQM0 B4LDP3 A0A1B0CJR8 B4PJ49 B4N3K5 B3M3S1 A0A0R1E182 A0A1W4USR4 B4KYL2 A0A0R1E087 A0A1I8PZF6 U5EY95 B4GRB6 Q2M0P8 A0A3B0K3X3 A0A0J9S020 B4IB57 Q9VNP7 A0A182S5D1 A0A2J7R742 A0A154NYY5 A0A0A1WPE0 A0A1L8EC89 A0A1L8ECN8 B3NEH0 A0A034WEG6 A0A1B6M6V6 A0A0K8WKD0 A0A1B6LT89 A0A0L0CRI8 E2AW33 A0A3F2YU02 A0A1A9WS78 W8BD52 A0A2M4CQZ9 B0WAU0 A0A2M4CPT0 A0A087Z982 A0A2M4CNC7 A0A182M0K0 Q16N01 A0A1A9UXV0 A0A1Q3F5L5 A0A1A9XBJ8 A0A1B0BAC4 A0A1A9Z3L8 A0A1B0FJ22 W5JHN2 A0A1L8EAK5 A0A1Y0AWR5 W5JED8 A0A1A9UMN4 A0A1B0GNR1 A0A1A9XXE3 A0A1A9ZJB6 A0A1B6MDT5 A0A088A576 A0A1B6M8U7 A0A182UW64 A0A182VTB2 A0A182UCU2 A0A1A9VZJ1 A0A2H8U154 A0A1B6D1Q2 A0A1B6DYI8 A0A1B0FN85 A0A2M4BHL2 A0A182R1G1 A0A1B0B0Z1 A0A182JIS5 V5GYR1
A0A212F0U1 A0A2A4IUH9 A0A026X573 A0A310STV1 A0A2W1BFK7 A0A2A4IX97 A0A2A3ERP8 A0A195ET83 A0A151I7E1 K7IVG0 E9IB29 A0A0M9ACZ5 A0A158NVX0 A0A151WSG4 F4X4X5 A0A067RRF9 A0A336LKG1 A0A0L7QXA2 A0A151HYS8 B4J3S2 A0A1I8MEN9 A0A1J1HMV0 A0A1Y1MQL8 A0A1Y1MQM0 B4LDP3 A0A1B0CJR8 B4PJ49 B4N3K5 B3M3S1 A0A0R1E182 A0A1W4USR4 B4KYL2 A0A0R1E087 A0A1I8PZF6 U5EY95 B4GRB6 Q2M0P8 A0A3B0K3X3 A0A0J9S020 B4IB57 Q9VNP7 A0A182S5D1 A0A2J7R742 A0A154NYY5 A0A0A1WPE0 A0A1L8EC89 A0A1L8ECN8 B3NEH0 A0A034WEG6 A0A1B6M6V6 A0A0K8WKD0 A0A1B6LT89 A0A0L0CRI8 E2AW33 A0A3F2YU02 A0A1A9WS78 W8BD52 A0A2M4CQZ9 B0WAU0 A0A2M4CPT0 A0A087Z982 A0A2M4CNC7 A0A182M0K0 Q16N01 A0A1A9UXV0 A0A1Q3F5L5 A0A1A9XBJ8 A0A1B0BAC4 A0A1A9Z3L8 A0A1B0FJ22 W5JHN2 A0A1L8EAK5 A0A1Y0AWR5 W5JED8 A0A1A9UMN4 A0A1B0GNR1 A0A1A9XXE3 A0A1A9ZJB6 A0A1B6MDT5 A0A088A576 A0A1B6M8U7 A0A182UW64 A0A182VTB2 A0A182UCU2 A0A1A9VZJ1 A0A2H8U154 A0A1B6D1Q2 A0A1B6DYI8 A0A1B0FN85 A0A2M4BHL2 A0A182R1G1 A0A1B0B0Z1 A0A182JIS5 V5GYR1
Pubmed
19121390
26354079
22118469
24508170
30249741
28756777
+ More
20075255 21282665 21347285 21719571 24845553 17994087 25315136 28004739 18057021 17550304 15632085 23185243 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 25348373 26108605 20798317 24495485 20920257 17510324 26449545 28341416 23761445
20075255 21282665 21347285 21719571 24845553 17994087 25315136 28004739 18057021 17550304 15632085 23185243 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 25348373 26108605 20798317 24495485 20920257 17510324 26449545 28341416 23761445
EMBL
BABH01032762
BABH01032759
BABH01032760
RSAL01000147
RVE45876.1
ODYU01005059
+ More
SOQ45566.1 KQ461196 KPJ06300.1 KQ459580 KPI99332.1 AGBW02011015 OWR47368.1 NWSH01006137 PCG63637.1 KK107020 QOIP01000006 EZA62579.1 RLU21141.1 KQ759893 OAD62068.1 KZ150080 PZC73889.1 NWSH01005355 PCG64279.1 KZ288191 PBC34473.1 KQ981993 KYN31122.1 KQ978426 KYM94033.1 GL762106 EFZ22227.1 KQ435687 KOX81346.1 ADTU01027569 ADTU01027570 ADTU01027571 KQ982782 KYQ50731.1 GL888679 EGI58548.1 KK852470 KDR23205.1 UFQS01000039 UFQT01000039 SSW98116.1 SSX18502.1 KQ414705 KOC63248.1 KQ976730 KYM76403.1 CH916366 EDV97303.1 CVRI01000011 CRK89275.1 GEZM01024123 JAV87953.1 GEZM01024124 JAV87952.1 CH940647 EDW70004.1 KRF84667.1 AJWK01014899 CM000159 EDW95573.1 KRK02523.1 KRK02524.1 KRK02525.1 KRK02528.1 KRK02529.1 CH964095 EDW79210.1 CH902618 EDV41402.1 KPU79093.1 KRK02526.1 CH933809 EDW18823.1 KRG06381.1 KRK02527.1 GANO01002139 JAB57732.1 CH479188 EDW40301.1 CH379069 EAL30882.1 KRT07734.1 OUUW01000012 SPP87392.1 CM002912 KMZ01252.1 CH480826 EDW44520.1 AE014296 AY094824 AAF51880.1 AAF51881.1 AAF51882.1 AAM11177.1 NEVH01006738 PNF36658.1 KQ434783 KZC04782.1 GBXI01013942 JAD00350.1 GFDG01002441 JAV16358.1 GFDG01002440 JAV16359.1 CH954178 EDV52805.1 KQS44446.1 GAKP01005863 GAKP01005862 JAC53089.1 GEBQ01008325 JAT31652.1 GDHF01007363 GDHF01000708 JAI44951.1 JAI51606.1 GEBQ01013138 JAT26839.1 JRES01000005 KNC34980.1 GL443213 EFN62455.1 GAMC01015489 GAMC01015488 GAMC01015487 GAMC01015486 GAMC01015485 GAMC01015484 JAB91066.1 GGFL01003140 MBW67318.1 DS231875 EDS41806.1 GGFL01003139 MBW67317.1 GGFL01002230 MBW66408.1 AXCM01003009 KM593906 KX289322 CH477843 AKN80692.1 ANI24619.1 EAT35727.1 GFDL01012184 JAV22861.1 JXJN01010916 CCAG010018371 ADMH02001479 ETN62400.1 GFDG01003196 JAV15603.1 KY921795 ART29384.1 ETN62401.1 AJVK01013542 AJVK01013543 AJVK01013544 GEBQ01005897 JAT34080.1 GEBQ01007703 JAT32274.1 GFXV01008115 MBW19920.1 GEDC01017684 JAS19614.1 GEDC01006564 JAS30734.1 CCAG010010124 GGFJ01003404 MBW52545.1 AXCN02002053 JXJN01006906 GALX01002883 JAB65583.1
SOQ45566.1 KQ461196 KPJ06300.1 KQ459580 KPI99332.1 AGBW02011015 OWR47368.1 NWSH01006137 PCG63637.1 KK107020 QOIP01000006 EZA62579.1 RLU21141.1 KQ759893 OAD62068.1 KZ150080 PZC73889.1 NWSH01005355 PCG64279.1 KZ288191 PBC34473.1 KQ981993 KYN31122.1 KQ978426 KYM94033.1 GL762106 EFZ22227.1 KQ435687 KOX81346.1 ADTU01027569 ADTU01027570 ADTU01027571 KQ982782 KYQ50731.1 GL888679 EGI58548.1 KK852470 KDR23205.1 UFQS01000039 UFQT01000039 SSW98116.1 SSX18502.1 KQ414705 KOC63248.1 KQ976730 KYM76403.1 CH916366 EDV97303.1 CVRI01000011 CRK89275.1 GEZM01024123 JAV87953.1 GEZM01024124 JAV87952.1 CH940647 EDW70004.1 KRF84667.1 AJWK01014899 CM000159 EDW95573.1 KRK02523.1 KRK02524.1 KRK02525.1 KRK02528.1 KRK02529.1 CH964095 EDW79210.1 CH902618 EDV41402.1 KPU79093.1 KRK02526.1 CH933809 EDW18823.1 KRG06381.1 KRK02527.1 GANO01002139 JAB57732.1 CH479188 EDW40301.1 CH379069 EAL30882.1 KRT07734.1 OUUW01000012 SPP87392.1 CM002912 KMZ01252.1 CH480826 EDW44520.1 AE014296 AY094824 AAF51880.1 AAF51881.1 AAF51882.1 AAM11177.1 NEVH01006738 PNF36658.1 KQ434783 KZC04782.1 GBXI01013942 JAD00350.1 GFDG01002441 JAV16358.1 GFDG01002440 JAV16359.1 CH954178 EDV52805.1 KQS44446.1 GAKP01005863 GAKP01005862 JAC53089.1 GEBQ01008325 JAT31652.1 GDHF01007363 GDHF01000708 JAI44951.1 JAI51606.1 GEBQ01013138 JAT26839.1 JRES01000005 KNC34980.1 GL443213 EFN62455.1 GAMC01015489 GAMC01015488 GAMC01015487 GAMC01015486 GAMC01015485 GAMC01015484 JAB91066.1 GGFL01003140 MBW67318.1 DS231875 EDS41806.1 GGFL01003139 MBW67317.1 GGFL01002230 MBW66408.1 AXCM01003009 KM593906 KX289322 CH477843 AKN80692.1 ANI24619.1 EAT35727.1 GFDL01012184 JAV22861.1 JXJN01010916 CCAG010018371 ADMH02001479 ETN62400.1 GFDG01003196 JAV15603.1 KY921795 ART29384.1 ETN62401.1 AJVK01013542 AJVK01013543 AJVK01013544 GEBQ01005897 JAT34080.1 GEBQ01007703 JAT32274.1 GFXV01008115 MBW19920.1 GEDC01017684 JAS19614.1 GEDC01006564 JAS30734.1 CCAG010010124 GGFJ01003404 MBW52545.1 AXCN02002053 JXJN01006906 GALX01002883 JAB65583.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000007151
UP000218220
+ More
UP000053097 UP000279307 UP000242457 UP000078541 UP000078542 UP000002358 UP000053105 UP000005205 UP000075809 UP000007755 UP000027135 UP000053825 UP000078540 UP000001070 UP000095301 UP000183832 UP000008792 UP000092461 UP000002282 UP000007798 UP000007801 UP000192221 UP000009192 UP000095300 UP000008744 UP000001819 UP000268350 UP000001292 UP000000803 UP000075900 UP000235965 UP000076502 UP000008711 UP000037069 UP000000311 UP000075881 UP000091820 UP000002320 UP000075883 UP000008820 UP000078200 UP000092443 UP000092460 UP000092445 UP000092444 UP000000673 UP000092462 UP000005203 UP000075903 UP000075920 UP000075902 UP000075886 UP000075880
UP000053097 UP000279307 UP000242457 UP000078541 UP000078542 UP000002358 UP000053105 UP000005205 UP000075809 UP000007755 UP000027135 UP000053825 UP000078540 UP000001070 UP000095301 UP000183832 UP000008792 UP000092461 UP000002282 UP000007798 UP000007801 UP000192221 UP000009192 UP000095300 UP000008744 UP000001819 UP000268350 UP000001292 UP000000803 UP000075900 UP000235965 UP000076502 UP000008711 UP000037069 UP000000311 UP000075881 UP000091820 UP000002320 UP000075883 UP000008820 UP000078200 UP000092443 UP000092460 UP000092445 UP000092444 UP000000673 UP000092462 UP000005203 UP000075903 UP000075920 UP000075902 UP000075886 UP000075880
ProteinModelPortal
H9JXK0
H9JXJ9
A0A3S2TH46
A0A2H1VXW1
A0A194QMV8
A0A194Q172
+ More
A0A212F0U1 A0A2A4IUH9 A0A026X573 A0A310STV1 A0A2W1BFK7 A0A2A4IX97 A0A2A3ERP8 A0A195ET83 A0A151I7E1 K7IVG0 E9IB29 A0A0M9ACZ5 A0A158NVX0 A0A151WSG4 F4X4X5 A0A067RRF9 A0A336LKG1 A0A0L7QXA2 A0A151HYS8 B4J3S2 A0A1I8MEN9 A0A1J1HMV0 A0A1Y1MQL8 A0A1Y1MQM0 B4LDP3 A0A1B0CJR8 B4PJ49 B4N3K5 B3M3S1 A0A0R1E182 A0A1W4USR4 B4KYL2 A0A0R1E087 A0A1I8PZF6 U5EY95 B4GRB6 Q2M0P8 A0A3B0K3X3 A0A0J9S020 B4IB57 Q9VNP7 A0A182S5D1 A0A2J7R742 A0A154NYY5 A0A0A1WPE0 A0A1L8EC89 A0A1L8ECN8 B3NEH0 A0A034WEG6 A0A1B6M6V6 A0A0K8WKD0 A0A1B6LT89 A0A0L0CRI8 E2AW33 A0A3F2YU02 A0A1A9WS78 W8BD52 A0A2M4CQZ9 B0WAU0 A0A2M4CPT0 A0A087Z982 A0A2M4CNC7 A0A182M0K0 Q16N01 A0A1A9UXV0 A0A1Q3F5L5 A0A1A9XBJ8 A0A1B0BAC4 A0A1A9Z3L8 A0A1B0FJ22 W5JHN2 A0A1L8EAK5 A0A1Y0AWR5 W5JED8 A0A1A9UMN4 A0A1B0GNR1 A0A1A9XXE3 A0A1A9ZJB6 A0A1B6MDT5 A0A088A576 A0A1B6M8U7 A0A182UW64 A0A182VTB2 A0A182UCU2 A0A1A9VZJ1 A0A2H8U154 A0A1B6D1Q2 A0A1B6DYI8 A0A1B0FN85 A0A2M4BHL2 A0A182R1G1 A0A1B0B0Z1 A0A182JIS5 V5GYR1
A0A212F0U1 A0A2A4IUH9 A0A026X573 A0A310STV1 A0A2W1BFK7 A0A2A4IX97 A0A2A3ERP8 A0A195ET83 A0A151I7E1 K7IVG0 E9IB29 A0A0M9ACZ5 A0A158NVX0 A0A151WSG4 F4X4X5 A0A067RRF9 A0A336LKG1 A0A0L7QXA2 A0A151HYS8 B4J3S2 A0A1I8MEN9 A0A1J1HMV0 A0A1Y1MQL8 A0A1Y1MQM0 B4LDP3 A0A1B0CJR8 B4PJ49 B4N3K5 B3M3S1 A0A0R1E182 A0A1W4USR4 B4KYL2 A0A0R1E087 A0A1I8PZF6 U5EY95 B4GRB6 Q2M0P8 A0A3B0K3X3 A0A0J9S020 B4IB57 Q9VNP7 A0A182S5D1 A0A2J7R742 A0A154NYY5 A0A0A1WPE0 A0A1L8EC89 A0A1L8ECN8 B3NEH0 A0A034WEG6 A0A1B6M6V6 A0A0K8WKD0 A0A1B6LT89 A0A0L0CRI8 E2AW33 A0A3F2YU02 A0A1A9WS78 W8BD52 A0A2M4CQZ9 B0WAU0 A0A2M4CPT0 A0A087Z982 A0A2M4CNC7 A0A182M0K0 Q16N01 A0A1A9UXV0 A0A1Q3F5L5 A0A1A9XBJ8 A0A1B0BAC4 A0A1A9Z3L8 A0A1B0FJ22 W5JHN2 A0A1L8EAK5 A0A1Y0AWR5 W5JED8 A0A1A9UMN4 A0A1B0GNR1 A0A1A9XXE3 A0A1A9ZJB6 A0A1B6MDT5 A0A088A576 A0A1B6M8U7 A0A182UW64 A0A182VTB2 A0A182UCU2 A0A1A9VZJ1 A0A2H8U154 A0A1B6D1Q2 A0A1B6DYI8 A0A1B0FN85 A0A2M4BHL2 A0A182R1G1 A0A1B0B0Z1 A0A182JIS5 V5GYR1
PDB
6F34
E-value=9.74167e-55,
Score=542
Ontologies
GO
Topology
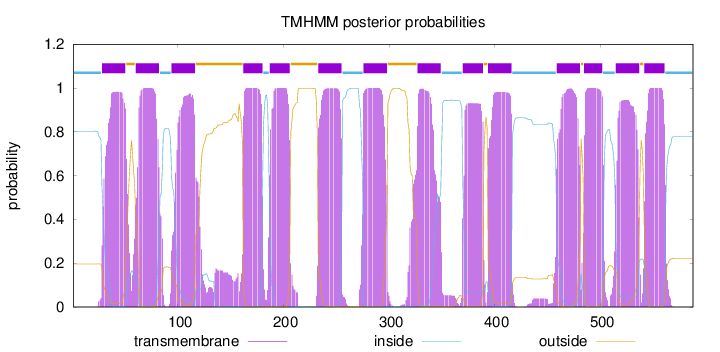
Length:
588
Number of predicted TMHs:
14
Exp number of AAs in TMHs:
301.01331
Exp number, first 60 AAs:
22.99846
Total prob of N-in:
0.80311
POSSIBLE N-term signal
sequence
inside
1 - 27
TMhelix
28 - 50
outside
51 - 59
TMhelix
60 - 82
inside
83 - 93
TMhelix
94 - 116
outside
117 - 161
TMhelix
162 - 180
inside
181 - 186
TMhelix
187 - 206
outside
207 - 232
TMhelix
233 - 255
inside
256 - 275
TMhelix
276 - 298
outside
299 - 326
TMhelix
327 - 349
inside
350 - 369
TMhelix
370 - 389
outside
390 - 393
TMhelix
394 - 416
inside
417 - 458
TMhelix
459 - 481
outside
482 - 484
TMhelix
485 - 502
inside
503 - 514
TMhelix
515 - 537
outside
538 - 541
TMhelix
542 - 561
inside
562 - 588
Population Genetic Test Statistics
Pi
205.553514
Theta
166.033235
Tajima's D
-1.226704
CLR
19.066437
CSRT
0.095645217739113
Interpretation
Uncertain
