Gene
KWMTBOMO04251
Pre Gene Modal
BGIBMGA014240
Annotation
PREDICTED:_cationic_amino_acid_transporter_2-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.848
Sequence
CDS
ATGGCTGGTGGGAAGTGGACGTTTTGCGACGTCGTGCGCCGGCGGAAGATTATACAGCCTCATCAGCTTGAAGCTGGAAATTTGAAAAGATGCCTGAGTCTTTGGGACGTGACAGCCCTTGGGGTGGGCAGTACTCTCGGAGCTGGTGTCTACGTCATTATCGGTTATGCAGCCTTCAAGTACGCAGGACCTTCAATTGTGCTCTCGTTCCTGATAGCTGGAGTTGCGGCTTTATTTTCAGGTTTGTGTTACGCTGAATTTGGAGCCAGAGTACCAAGAGCTGGTTCAGCATATGTTTACAGCTACGTGGCCATCGGGGAGGTGGTGGCTTTCTTTATTGGTTGGTGCAATATTCTTGAAGCAGCGATGGGCGCAGCGAGCTTGGCCCGCGGTCTCAGCATGTACGTGGACGGGATGTGCAACAATTCAATATTAGAATGGGCAACAGCTACAATGCCGATCTCTTCCTCATTCTTGTCACCGTACTTTGATCTGCTGGCGTTTGCATTTGTTCTTGTCATTGGAGTACTGCTATCAGTGGGTGGTCGAGAGTCGTCCGCCGTGAACATCGCGTTTGTCTTGCTGAACTTGTTAGTGATTATACTTGTTGTCATCGTTGGAGCTATCAATGCTGATTCAGCTAACTGGAGTATCCCATCCACGGACGTTCCGCAAGGTAGTGGCTCTGGAGGATTCTTCCCATACGGCGTATGGGGCACACTGAAGGGCGCTGCGATTTGCTTTTATGGTTTTGTTGGATTCGATACCATTAACGCTGCGGCCGAAGAGGTTCGAAAACCTCAAGAAACCATTCCAATGGTAATATTGATAGTTCTGCTTACTGCATTTGTTTGTTATTGCGGAATTTCTATTGTTTTGACAATGATGGTGCCATACTATCTGCAGAACTCCACCGATGCTGTTGGGTCAGCGTTCATTTTCATCGGATGGGAGTGGATGAAGTGGGTGGTATTTGTTGGTGCCTTCGCTGGGATCCTTGCCAGTTTATTTGGGGCCTTGTTGCCTCTCCCGCGGCTGTTGTACGCGATGTCCTCAGACGGCTTATTGGTCAGTTGGTTTTCGAAGTTAACGTCGTCGCGCAAGTCTCCTGTTTTTGCCACGATTTCGTCAGCTGCTGTTGTTGCCATACTTGCTGGAATTTTGGAACTAGAACAACTCATACTGATGCTTTGTATTGGAACGCTGCTCTCTTACACCGTAGTGGCTGTCTGTGTGATCGTATTAAGGTTTCGTTCAGAATACGCTCCTCAATCGTCTCCAGGGTTCTTCAAGGAGGTATTCGGCTGTGGCTTCAGGCTCGCCACACGTAGCACCGCCAGGATCGTTTACGCAGTTTTGTTCCTCTTTATAAGCGTATGTGTCTCTATGGCACTTATCTTGGCTCACGTGGAGAGGCCTCTCATACCACTTCTGGTACTACACGTGCTGGCTTTGATGCTGGTCATTATTATGTCGCTTCAGCCTAAAGCGAAGGAGGAGGTAGCTTTTAAGACTCCTCTGGTGCCTCTGATTCCCTGTCTGAGCATATACGTGAACGTGCACTTGATGGCCCTCATCAAACTTCAGACTTGGATTAGAGTCCTGATTTGGCTTGCTATAGGTATACCAGTCTACTTATTAGGATTATGTTGTTTCAAGAAGAAAGACAAAGAAGAAAAGGATATAGTTATGCCACACTCAGATGAGAATGGGAAACCACTACCTGTACAAATAGTAGTAGAATCACCTACGCCGCCTGATAGTATTAATAGAATTGGTAGCCACGGGGACAATACTATACTGGAAGAGAACGAACAAAGTAGCGATCAAATACGTAATGTCAGAAATGAAACATTCAATATCGAGGAAATCAAACATCAAGTGATTATTGAGAATAATGAAGAAAAGGAAGCTAAAATAATAGACCTGTTAGATCAAGTCCTGCAAGCTGAAGAAGAAACATACGGAGAAGCTATCAGTCTGAAAGACCAAAATATAGAAGAAGCTCCTGATGTCAAAGAAGTTATCCACAGAAAATCACTAGGCGAGCTTTCTGACGCAGGGTCTGATGTATCATCAGGTAATCAAGTATTATCTAAATATGATGTAATAGCGCAGGTCCACAGAGAGGATCTGCCTAAGGTAACTGAAGAAGAAGAAGAAAAAAGAGATGATGACGATGATCTAGAACAAGAAACAGAACAACACGAGCAAATCACTGCTTTTAATGACAGTGAGACCAATTCAAGGACCGATGAATCTGGATATTCTGACACGTTAGATAGAAATCCTTTCAGTGAGTCTCTTGAAGAGAAAGAGGATAGGGATGAAGAAGAGGCTCCTAATATACCAGTGCCACCGCCATTAGATGAAAATTTTTTTAGAAGTCCAAATTTTAAGAAATCTTATACAATTTCATCGAGACCTTTAAAAAAAGAAATGTCCGAAGAAGCAGAAGAGATTCCAAGACAAAGCGTTCAGTCGAATAACTCTCATAGCGATGACAATATTACGTTTGGTAGTGATAGACAAAAACATTTCATGAGTAAGCTAAACGATATATTTCAAAACAAGATAACGAATAACCAGGATGATGATGATGAAGAACCAAGAAAAAGATCACAGTCGACTGGAAATGCACCTGAAGACAACACTTTCACCGTAAGTCGTCCGTCTATATTCCTAGATTTGAAAAAGGAGATTGTCTCTCGTGAAGTGGCTCAGAATTTGCGACCTGTCAATCCAGACGAGACGAAAACCGAAGAAGAATCAACAGAAGAAGACAAACAATTGACAAGAGCAGATTTAAAATCTAAGTTAGAAAATATATTCGCTGCGGGCGGCCCTCAACTATTAAAACCTAGGCTGATGAAATCTAATCCTCCTACACCTGAAGAAGCATATCAAACTGATACATCGAGCACAGAAAGTATTGCCAAATTAGCGAAGCTGGACAAAAATGATACTTTAAAACGTCAAAAAGCTAAGTTCGGTGAAGTATTGAATTCCTTTAGGCTCAGTTTTAACAAAGACGATGCAGTTTGA
Protein
MAGGKWTFCDVVRRRKIIQPHQLEAGNLKRCLSLWDVTALGVGSTLGAGVYVIIGYAAFKYAGPSIVLSFLIAGVAALFSGLCYAEFGARVPRAGSAYVYSYVAIGEVVAFFIGWCNILEAAMGAASLARGLSMYVDGMCNNSILEWATATMPISSSFLSPYFDLLAFAFVLVIGVLLSVGGRESSAVNIAFVLLNLLVIILVVIVGAINADSANWSIPSTDVPQGSGSGGFFPYGVWGTLKGAAICFYGFVGFDTINAAAEEVRKPQETIPMVILIVLLTAFVCYCGISIVLTMMVPYYLQNSTDAVGSAFIFIGWEWMKWVVFVGAFAGILASLFGALLPLPRLLYAMSSDGLLVSWFSKLTSSRKSPVFATISSAAVVAILAGILELEQLILMLCIGTLLSYTVVAVCVIVLRFRSEYAPQSSPGFFKEVFGCGFRLATRSTARIVYAVLFLFISVCVSMALILAHVERPLIPLLVLHVLALMLVIIMSLQPKAKEEVAFKTPLVPLIPCLSIYVNVHLMALIKLQTWIRVLIWLAIGIPVYLLGLCCFKKKDKEEKDIVMPHSDENGKPLPVQIVVESPTPPDSINRIGSHGDNTILEENEQSSDQIRNVRNETFNIEEIKHQVIIENNEEKEAKIIDLLDQVLQAEEETYGEAISLKDQNIEEAPDVKEVIHRKSLGELSDAGSDVSSGNQVLSKYDVIAQVHREDLPKVTEEEEEKRDDDDDLEQETEQHEQITAFNDSETNSRTDESGYSDTLDRNPFSESLEEKEDRDEEEAPNIPVPPPLDENFFRSPNFKKSYTISSRPLKKEMSEEAEEIPRQSVQSNNSHSDDNITFGSDRQKHFMSKLNDIFQNKITNNQDDDDEEPRKRSQSTGNAPEDNTFTVSRPSIFLDLKKEIVSREVAQNLRPVNPDETKTEEESTEEDKQLTRADLKSKLENIFAAGGPQLLKPRLMKSNPPTPEEAYQTDTSSTESIAKLAKLDKNDTLKRQKAKFGEVLNSFRLSFNKDDAV
Summary
Uniprot
EMBL
Proteomes
PRIDE
ProteinModelPortal
PDB
6F34
E-value=2.16474e-56,
Score=558
Ontologies
GO
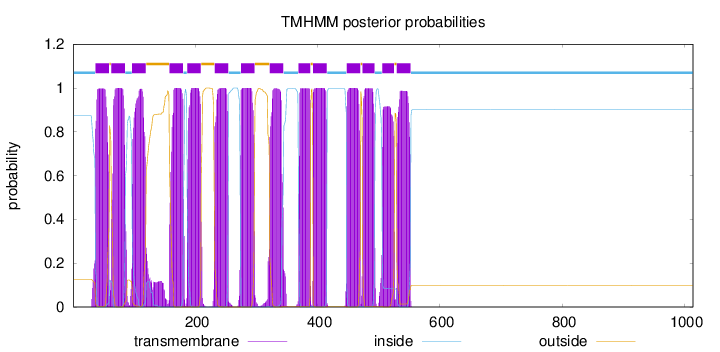
Topology
Length:
1014
Number of predicted TMHs:
14
Exp number of AAs in TMHs:
306.57073
Exp number, first 60 AAs:
22.24583
Total prob of N-in:
0.87347
POSSIBLE N-term signal
sequence
inside
1 - 36
TMhelix
37 - 59
outside
60 - 62
TMhelix
63 - 85
inside
86 - 96
TMhelix
97 - 119
outside
120 - 157
TMhelix
158 - 180
inside
181 - 186
TMhelix
187 - 209
outside
210 - 231
TMhelix
232 - 254
inside
255 - 274
TMhelix
275 - 297
outside
298 - 321
TMhelix
322 - 344
inside
345 - 368
TMhelix
369 - 388
outside
389 - 392
TMhelix
393 - 415
inside
416 - 447
TMhelix
448 - 470
outside
471 - 473
TMhelix
474 - 493
inside
494 - 505
TMhelix
506 - 525
outside
526 - 529
TMhelix
530 - 552
inside
553 - 1014
Population Genetic Test Statistics
Pi
236.286151
Theta
179.950848
Tajima's D
0.653784
CLR
0.531381
CSRT
0.567121643917804
Interpretation
Uncertain
