Gene
KWMTBOMO04243
Pre Gene Modal
BGIBMGA014261
Annotation
PREDICTED:_metabotropic_glutamate_receptor_2-like_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.798
Sequence
CDS
ATGCGGCCGGCGAGTGGCGCTGATTTACTGCGCTACCTAAGGAAAGTCAACTTTACTGGTGTGAGCGGTGACGAGTTCCACTTCGACAGCAATGGCGACGGTCCAGCCAGGTATAACATCCTGCACTTCAAGCAAGTCTCACGAGGAACATACAGCTGGGTTAACGTTGGCCAGTACTTGGATGGAGAACTTCAACTACATATCAAGGACATTCAGTTCAAGTGGGGAGAAAGACATCCACCGGAGTCAGTGTGCAGCGCGGAGTGCGAACTCGGACAAGCCAAGCAGTACGTGGAGGGCGAGAGTTGCTGCTGGCACTGCTTCAACTGCACTCAGTACGAGATCCGGTCTCCGTACGTGGAGACAGCATGTATGGTGTGTCCGCGCGGTACGTTACCGGATCTAACCAGGACGCAATGTCGACCGATCCCTGAAGCGTACTTAAGACCTGACTCCGCGTGGGCGATCGGCGCTATGTCGTTCTCATCTGTCGGAATATTACTGACTGCATTTGTCTGCGGAGTGTGGGTTAGACACAGTTCGACTCCAGTAGTGCGTGCGAGCGGGAGGGAGCTATCCTACGTCCTGCTCGCCGGTATCCTCATGTGCTATCTCGTTACATTCGCCCTGGTGTTCAGGCCCACGGACATACTTTGCTCTATTCAAAGATTCGGCACCGGTTTCTGCTTTACGGTCATCTACGCCGCTCTTCTCACCAAGACAAACAGGATCTCCAGGATATTCAATGCCAGTAAACATTCAGCAAAAAGACCTATCCTCATATCTCCAACTTCCCAACTAGCTATATGCGCTGGACTTGTGTCCATTCAGGTACTTATAGTGGTAGTCTGGATGATAGTGGCCCCAGCCAGAGCGATGTTCCACTACCCGACCAGAGAAGACAACATGCTAGTTTGCGACTCCTACGTAGATGCTTCATACATGATAGCGTTTTTCTATCCCATCGTACTAATTCTCATCTGTACTGTCTACGCCGTTCTCACCAGGAAGATACCGGAGGCTTTCAACGAGAGCAAGCACATTGGTTTTACAATGTACACCACCTGCGTCATCTGGTTGGCATTTGTGCCGTTGTACTTCGGTACCGCGAGCCACGTGCCTTTGAGGGTCACCAGCATGGCGGTCACCATCTCCCTGAGCGCCAGCGTCACGCTCATCTGCTTGTTCTCGCCAAAGCTGTATATAATACTCATCCGTCCGGAGAGGAACGTGCGTCAAAGCATCATGCCGGTGAGGTATAGCCGGAAGCCGCCTCCGCCGCATCCGCTAAACATCCAAGTCAAAAAACCACCGTGCGCCGACAAAGAGACGCAAACGGATCCGGCCGTGTTCAAGTTGTCCAACGGACAAGTCGTCAACGACACGACACGGAAAGAAGAGAAGATAAACAAAGATGAAAAAGCAAAAGAAACAAAACAATGTAACAACGTAAATAATGTTAGCGATTCGAAGAACACGCAGACTGAAAAGACCGCCTCCTTATAG
Protein
MRPASGADLLRYLRKVNFTGVSGDEFHFDSNGDGPARYNILHFKQVSRGTYSWVNVGQYLDGELQLHIKDIQFKWGERHPPESVCSAECELGQAKQYVEGESCCWHCFNCTQYEIRSPYVETACMVCPRGTLPDLTRTQCRPIPEAYLRPDSAWAIGAMSFSSVGILLTAFVCGVWVRHSSTPVVRASGRELSYVLLAGILMCYLVTFALVFRPTDILCSIQRFGTGFCFTVIYAALLTKTNRISRIFNASKHSAKRPILISPTSQLAICAGLVSIQVLIVVVWMIVAPARAMFHYPTREDNMLVCDSYVDASYMIAFFYPIVLILICTVYAVLTRKIPEAFNESKHIGFTMYTTCVIWLAFVPLYFGTASHVPLRVTSMAVTISLSASVTLICLFSPKLYIILIRPERNVRQSIMPVRYSRKPPPPHPLNIQVKKPPCADKETQTDPAVFKLSNGQVVNDTTRKEEKINKDEKAKETKQCNNVNNVSDSKNTQTEKTASL
Summary
Similarity
Belongs to the G-protein coupled receptor 3 family.
Uniprot
A0A2W1BFP1
A0A2H1VDD3
A0A194RGF0
A0A194Q8J6
A0A2A4J705
A0A2W1BGB3
+ More
A0A2A4JI84 A0A0P9ACD0 A0A0N0BDM9 A0A154PQC1 A0A1A9W8Y4 A0A0L7R3U0 B3MFX4 A0A0J9TWB7 A0A0K8U462 A0A0R1DM99 A0A1I8M0J9 A0A0Q5WKX7 A0A088AQD5 A0A2A3E9Y4 Q75QW6 A0A0B4KF15 A0A0R3NL13 B4LLC1 A0A0J9R8A8 A0A3B0IZT4 A0A0K8VB57 A1Z7F4 B7YZT0 B4P2U6 B4HRY5 A0A0L0CQG5 Q70GQ8 A1Z7F5 B3N8N5 W8BYX6 A0A1W4UJR8 B4GDF5 B4J5X5 B5DZA5 A1Z7F6 B4MQ18 B4KP51 A0A1Y1LYJ4 K7J0X5 A0A0M3QUN3 A0A232EQZ0 A0A195D7C2 A0A151WWG1 A0A2J7RRI9 A0A212FEA2 A0A1B0C9E3 E0VG09 A0A195DLK5 A0A2J7RRH8 A0A195B3M6 A0A1J1J939 A0A067R1J1 A0A1B0GBF7 A0A182RUV7 F4WBG0 A0A195FTU1 E2C1A5 A0A182NLU4 A0A182HPF8 A0A158P133 Q7Q9V1 A0A182X0Z4 A0A182YJ34 A0A212EHW5 A0A084WKZ5 A0A1W7R6Q5 A0A182S9N9 A0A336KSK2 A0A182PRF3 T1HKT5 A0A0A9YU67 A0A0A9XNL6 A0A0A9Z1S2 J9JLK4 E2AUR2 A0A2S2PI57 A0A026VXM2 A0A2H8TP95 T1I399 A0A0L7LCG4 A0A139WCP2 A0A139WCD7 Q16UR9 T1HFM1 B4NSA6 A0A0P5L347 A0A0P5QPR3 A0A0P5P337 A0A0P5NWW9 A0A0P5PG09 A0A0P6EYN9 A0A0P5MTW6 A0A0N8BUT1
A0A2A4JI84 A0A0P9ACD0 A0A0N0BDM9 A0A154PQC1 A0A1A9W8Y4 A0A0L7R3U0 B3MFX4 A0A0J9TWB7 A0A0K8U462 A0A0R1DM99 A0A1I8M0J9 A0A0Q5WKX7 A0A088AQD5 A0A2A3E9Y4 Q75QW6 A0A0B4KF15 A0A0R3NL13 B4LLC1 A0A0J9R8A8 A0A3B0IZT4 A0A0K8VB57 A1Z7F4 B7YZT0 B4P2U6 B4HRY5 A0A0L0CQG5 Q70GQ8 A1Z7F5 B3N8N5 W8BYX6 A0A1W4UJR8 B4GDF5 B4J5X5 B5DZA5 A1Z7F6 B4MQ18 B4KP51 A0A1Y1LYJ4 K7J0X5 A0A0M3QUN3 A0A232EQZ0 A0A195D7C2 A0A151WWG1 A0A2J7RRI9 A0A212FEA2 A0A1B0C9E3 E0VG09 A0A195DLK5 A0A2J7RRH8 A0A195B3M6 A0A1J1J939 A0A067R1J1 A0A1B0GBF7 A0A182RUV7 F4WBG0 A0A195FTU1 E2C1A5 A0A182NLU4 A0A182HPF8 A0A158P133 Q7Q9V1 A0A182X0Z4 A0A182YJ34 A0A212EHW5 A0A084WKZ5 A0A1W7R6Q5 A0A182S9N9 A0A336KSK2 A0A182PRF3 T1HKT5 A0A0A9YU67 A0A0A9XNL6 A0A0A9Z1S2 J9JLK4 E2AUR2 A0A2S2PI57 A0A026VXM2 A0A2H8TP95 T1I399 A0A0L7LCG4 A0A139WCP2 A0A139WCD7 Q16UR9 T1HFM1 B4NSA6 A0A0P5L347 A0A0P5QPR3 A0A0P5P337 A0A0P5NWW9 A0A0P5PG09 A0A0P6EYN9 A0A0P5MTW6 A0A0N8BUT1
Pubmed
28756777
26354079
17994087
22936249
17550304
25315136
+ More
15050695 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 26108605 14660580 24495485 26109357 26109356 28004739 20075255 28648823 22118469 20566863 24845553 21719571 20798317 21347285 12364791 25244985 24438588 25401762 26823975 24508170 26227816 18362917 19820115 17510324
15050695 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 26108605 14660580 24495485 26109357 26109356 28004739 20075255 28648823 22118469 20566863 24845553 21719571 20798317 21347285 12364791 25244985 24438588 25401762 26823975 24508170 26227816 18362917 19820115 17510324
EMBL
KZ150080
PZC73882.1
ODYU01001945
SOQ38848.1
KQ460211
KPJ16524.1
+ More
KQ459302 KPJ01848.1 NWSH01002590 PCG67907.1 KZ150487 PZC70703.1 NWSH01001473 PCG71140.1 CH902619 KPU75778.1 KQ435855 KOX70662.1 KQ435036 KZC14082.1 KQ414661 KOC65555.1 EDV35656.2 CM002911 KMY92273.1 KMY92274.1 KMY92275.1 GDHF01030797 JAI21517.1 CM000157 KRJ98364.1 CH954177 KQS70875.1 KZ288317 PBC28294.1 AB161182 BAD08344.1 AE013599 AGB93339.1 CM000071 KRT01640.1 CH940648 EDW60858.2 KMY92276.1 OUUW01000001 SPP73687.1 GDHF01016218 JAI36096.1 AAF59081.3 ACL83073.1 EDW89357.2 CH480816 EDW46948.1 JRES01000171 KNC33679.1 AJ581534 CAE46392.1 AAF59082.3 EDV59512.1 GAMC01002043 JAC04513.1 CH479181 EDW31631.1 CH916367 EDW00818.1 EDY68791.2 BT033068 AAS64894.1 ACE82591.1 CH963849 EDW74207.1 CH933808 EDW10117.2 GEZM01047799 JAV76746.1 CP012524 ALC40933.1 NNAY01002683 OXU20784.1 KQ976749 KYN08785.1 KQ982691 KYQ52223.1 NEVH01000606 PNF43434.1 AGBW02008952 OWR52095.1 AJWK01002248 DS235129 EEB12315.1 KQ980765 KYN13374.1 PNF43433.1 KQ976641 KYM78799.1 CVRI01000074 CRL08038.1 KK852776 KDR16788.1 CCAG010014001 CCAG010014002 GL888063 EGI68439.1 KQ981272 KYN43846.1 GL451914 EFN78267.1 APCN01002995 ADTU01006103 AAAB01008900 EAA09380.4 AGBW02014793 OWR41061.1 ATLV01024163 KE525350 KFB50889.1 GEHC01000817 JAV46828.1 UFQS01000437 UFQT01000437 SSX03930.1 SSX24295.1 ACPB03017251 ACPB03017252 ACPB03017253 ACPB03017254 GBHO01007850 GDHC01013000 JAG35754.1 JAQ05629.1 GBHO01035356 GBHO01035354 GBHO01030107 GBHO01024922 GBHO01024919 GBHO01024918 GDHC01019174 JAG08248.1 JAG08250.1 JAG13497.1 JAG18682.1 JAG18685.1 JAG18686.1 JAP99454.1 GBHO01007849 GDHC01001086 JAG35755.1 JAQ17543.1 ABLF02023192 GL442888 EFN62787.1 GGMR01016299 MBY28918.1 KK107623 EZA48425.1 GFXV01003936 MBW15741.1 ACPB03010517 JTDY01001683 KOB73168.1 KQ971367 KYB25635.1 KYB25638.1 CH477615 EAT38269.2 ACPB03010165 ACPB03010166 CH982034 EDX15484.1 GDIQ01176096 JAK75629.1 GDIQ01111270 JAL40456.1 GDIQ01155161 JAK96564.1 GDIQ01136570 JAL15156.1 GDIQ01139071 JAL12655.1 GDIQ01057440 JAN37297.1 GDIQ01159362 JAK92363.1 GDIQ01142811 GDIQ01142809 JAL08915.1
KQ459302 KPJ01848.1 NWSH01002590 PCG67907.1 KZ150487 PZC70703.1 NWSH01001473 PCG71140.1 CH902619 KPU75778.1 KQ435855 KOX70662.1 KQ435036 KZC14082.1 KQ414661 KOC65555.1 EDV35656.2 CM002911 KMY92273.1 KMY92274.1 KMY92275.1 GDHF01030797 JAI21517.1 CM000157 KRJ98364.1 CH954177 KQS70875.1 KZ288317 PBC28294.1 AB161182 BAD08344.1 AE013599 AGB93339.1 CM000071 KRT01640.1 CH940648 EDW60858.2 KMY92276.1 OUUW01000001 SPP73687.1 GDHF01016218 JAI36096.1 AAF59081.3 ACL83073.1 EDW89357.2 CH480816 EDW46948.1 JRES01000171 KNC33679.1 AJ581534 CAE46392.1 AAF59082.3 EDV59512.1 GAMC01002043 JAC04513.1 CH479181 EDW31631.1 CH916367 EDW00818.1 EDY68791.2 BT033068 AAS64894.1 ACE82591.1 CH963849 EDW74207.1 CH933808 EDW10117.2 GEZM01047799 JAV76746.1 CP012524 ALC40933.1 NNAY01002683 OXU20784.1 KQ976749 KYN08785.1 KQ982691 KYQ52223.1 NEVH01000606 PNF43434.1 AGBW02008952 OWR52095.1 AJWK01002248 DS235129 EEB12315.1 KQ980765 KYN13374.1 PNF43433.1 KQ976641 KYM78799.1 CVRI01000074 CRL08038.1 KK852776 KDR16788.1 CCAG010014001 CCAG010014002 GL888063 EGI68439.1 KQ981272 KYN43846.1 GL451914 EFN78267.1 APCN01002995 ADTU01006103 AAAB01008900 EAA09380.4 AGBW02014793 OWR41061.1 ATLV01024163 KE525350 KFB50889.1 GEHC01000817 JAV46828.1 UFQS01000437 UFQT01000437 SSX03930.1 SSX24295.1 ACPB03017251 ACPB03017252 ACPB03017253 ACPB03017254 GBHO01007850 GDHC01013000 JAG35754.1 JAQ05629.1 GBHO01035356 GBHO01035354 GBHO01030107 GBHO01024922 GBHO01024919 GBHO01024918 GDHC01019174 JAG08248.1 JAG08250.1 JAG13497.1 JAG18682.1 JAG18685.1 JAG18686.1 JAP99454.1 GBHO01007849 GDHC01001086 JAG35755.1 JAQ17543.1 ABLF02023192 GL442888 EFN62787.1 GGMR01016299 MBY28918.1 KK107623 EZA48425.1 GFXV01003936 MBW15741.1 ACPB03010517 JTDY01001683 KOB73168.1 KQ971367 KYB25635.1 KYB25638.1 CH477615 EAT38269.2 ACPB03010165 ACPB03010166 CH982034 EDX15484.1 GDIQ01176096 JAK75629.1 GDIQ01111270 JAL40456.1 GDIQ01155161 JAK96564.1 GDIQ01136570 JAL15156.1 GDIQ01139071 JAL12655.1 GDIQ01057440 JAN37297.1 GDIQ01159362 JAK92363.1 GDIQ01142811 GDIQ01142809 JAL08915.1
Proteomes
UP000053240
UP000053268
UP000218220
UP000007801
UP000053105
UP000076502
+ More
UP000091820 UP000053825 UP000002282 UP000095301 UP000008711 UP000005203 UP000242457 UP000000803 UP000001819 UP000008792 UP000268350 UP000001292 UP000037069 UP000192221 UP000008744 UP000001070 UP000007798 UP000009192 UP000002358 UP000092553 UP000215335 UP000078542 UP000075809 UP000235965 UP000007151 UP000092461 UP000009046 UP000078492 UP000078540 UP000183832 UP000027135 UP000092444 UP000075900 UP000007755 UP000078541 UP000008237 UP000075884 UP000075840 UP000005205 UP000007062 UP000076407 UP000076408 UP000030765 UP000075901 UP000075885 UP000015103 UP000007819 UP000000311 UP000053097 UP000037510 UP000007266 UP000008820 UP000000304
UP000091820 UP000053825 UP000002282 UP000095301 UP000008711 UP000005203 UP000242457 UP000000803 UP000001819 UP000008792 UP000268350 UP000001292 UP000037069 UP000192221 UP000008744 UP000001070 UP000007798 UP000009192 UP000002358 UP000092553 UP000215335 UP000078542 UP000075809 UP000235965 UP000007151 UP000092461 UP000009046 UP000078492 UP000078540 UP000183832 UP000027135 UP000092444 UP000075900 UP000007755 UP000078541 UP000008237 UP000075884 UP000075840 UP000005205 UP000007062 UP000076407 UP000076408 UP000030765 UP000075901 UP000075885 UP000015103 UP000007819 UP000000311 UP000053097 UP000037510 UP000007266 UP000008820 UP000000304
Pfam
Interpro
IPR000162
GPCR_3_mtglu_rcpt
+ More
IPR001828 ANF_lig-bd_rcpt
IPR017978 GPCR_3_C
IPR038550 GPCR_3_9-Cys_sf
IPR017979 GPCR_3_CS
IPR028082 Peripla_BP_I
IPR011500 GPCR_3_9-Cys_dom
IPR000337 GPCR_3
IPR029030 Caspase-like_dom_sf
IPR000330 SNF2_N
IPR027417 P-loop_NTPase
IPR038718 SNF2-like_sf
IPR014001 Helicase_ATP-bd
IPR020838 DBINO
IPR001650 Helicase_C
IPR009030 Growth_fac_rcpt_cys_sf
IPR029058 AB_hydrolase
IPR006693 AB_hydrolase_lipase
IPR001828 ANF_lig-bd_rcpt
IPR017978 GPCR_3_C
IPR038550 GPCR_3_9-Cys_sf
IPR017979 GPCR_3_CS
IPR028082 Peripla_BP_I
IPR011500 GPCR_3_9-Cys_dom
IPR000337 GPCR_3
IPR029030 Caspase-like_dom_sf
IPR000330 SNF2_N
IPR027417 P-loop_NTPase
IPR038718 SNF2-like_sf
IPR014001 Helicase_ATP-bd
IPR020838 DBINO
IPR001650 Helicase_C
IPR009030 Growth_fac_rcpt_cys_sf
IPR029058 AB_hydrolase
IPR006693 AB_hydrolase_lipase
SUPFAM
Gene 3D
CDD
ProteinModelPortal
A0A2W1BFP1
A0A2H1VDD3
A0A194RGF0
A0A194Q8J6
A0A2A4J705
A0A2W1BGB3
+ More
A0A2A4JI84 A0A0P9ACD0 A0A0N0BDM9 A0A154PQC1 A0A1A9W8Y4 A0A0L7R3U0 B3MFX4 A0A0J9TWB7 A0A0K8U462 A0A0R1DM99 A0A1I8M0J9 A0A0Q5WKX7 A0A088AQD5 A0A2A3E9Y4 Q75QW6 A0A0B4KF15 A0A0R3NL13 B4LLC1 A0A0J9R8A8 A0A3B0IZT4 A0A0K8VB57 A1Z7F4 B7YZT0 B4P2U6 B4HRY5 A0A0L0CQG5 Q70GQ8 A1Z7F5 B3N8N5 W8BYX6 A0A1W4UJR8 B4GDF5 B4J5X5 B5DZA5 A1Z7F6 B4MQ18 B4KP51 A0A1Y1LYJ4 K7J0X5 A0A0M3QUN3 A0A232EQZ0 A0A195D7C2 A0A151WWG1 A0A2J7RRI9 A0A212FEA2 A0A1B0C9E3 E0VG09 A0A195DLK5 A0A2J7RRH8 A0A195B3M6 A0A1J1J939 A0A067R1J1 A0A1B0GBF7 A0A182RUV7 F4WBG0 A0A195FTU1 E2C1A5 A0A182NLU4 A0A182HPF8 A0A158P133 Q7Q9V1 A0A182X0Z4 A0A182YJ34 A0A212EHW5 A0A084WKZ5 A0A1W7R6Q5 A0A182S9N9 A0A336KSK2 A0A182PRF3 T1HKT5 A0A0A9YU67 A0A0A9XNL6 A0A0A9Z1S2 J9JLK4 E2AUR2 A0A2S2PI57 A0A026VXM2 A0A2H8TP95 T1I399 A0A0L7LCG4 A0A139WCP2 A0A139WCD7 Q16UR9 T1HFM1 B4NSA6 A0A0P5L347 A0A0P5QPR3 A0A0P5P337 A0A0P5NWW9 A0A0P5PG09 A0A0P6EYN9 A0A0P5MTW6 A0A0N8BUT1
A0A2A4JI84 A0A0P9ACD0 A0A0N0BDM9 A0A154PQC1 A0A1A9W8Y4 A0A0L7R3U0 B3MFX4 A0A0J9TWB7 A0A0K8U462 A0A0R1DM99 A0A1I8M0J9 A0A0Q5WKX7 A0A088AQD5 A0A2A3E9Y4 Q75QW6 A0A0B4KF15 A0A0R3NL13 B4LLC1 A0A0J9R8A8 A0A3B0IZT4 A0A0K8VB57 A1Z7F4 B7YZT0 B4P2U6 B4HRY5 A0A0L0CQG5 Q70GQ8 A1Z7F5 B3N8N5 W8BYX6 A0A1W4UJR8 B4GDF5 B4J5X5 B5DZA5 A1Z7F6 B4MQ18 B4KP51 A0A1Y1LYJ4 K7J0X5 A0A0M3QUN3 A0A232EQZ0 A0A195D7C2 A0A151WWG1 A0A2J7RRI9 A0A212FEA2 A0A1B0C9E3 E0VG09 A0A195DLK5 A0A2J7RRH8 A0A195B3M6 A0A1J1J939 A0A067R1J1 A0A1B0GBF7 A0A182RUV7 F4WBG0 A0A195FTU1 E2C1A5 A0A182NLU4 A0A182HPF8 A0A158P133 Q7Q9V1 A0A182X0Z4 A0A182YJ34 A0A212EHW5 A0A084WKZ5 A0A1W7R6Q5 A0A182S9N9 A0A336KSK2 A0A182PRF3 T1HKT5 A0A0A9YU67 A0A0A9XNL6 A0A0A9Z1S2 J9JLK4 E2AUR2 A0A2S2PI57 A0A026VXM2 A0A2H8TP95 T1I399 A0A0L7LCG4 A0A139WCP2 A0A139WCD7 Q16UR9 T1HFM1 B4NSA6 A0A0P5L347 A0A0P5QPR3 A0A0P5P337 A0A0P5NWW9 A0A0P5PG09 A0A0P6EYN9 A0A0P5MTW6 A0A0N8BUT1
PDB
6N52
E-value=3.63127e-93,
Score=872
Ontologies
GO
Topology
Subcellular location
Cell membrane
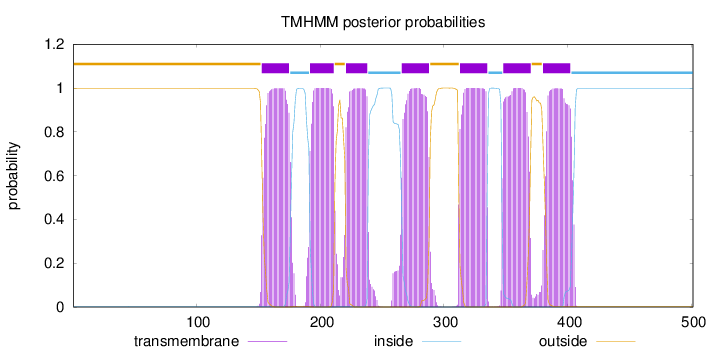
Length:
501
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
153.86702
Exp number, first 60 AAs:
0.00042
Total prob of N-in:
0.00264
outside
1 - 152
TMhelix
153 - 175
inside
176 - 191
TMhelix
192 - 211
outside
212 - 220
TMhelix
221 - 238
inside
239 - 265
TMhelix
266 - 288
outside
289 - 312
TMhelix
313 - 335
inside
336 - 347
TMhelix
348 - 370
outside
371 - 379
TMhelix
380 - 402
inside
403 - 501
Population Genetic Test Statistics
Pi
192.928262
Theta
149.457627
Tajima's D
1.388237
CLR
0.351538
CSRT
0.761661916904155
Interpretation
Uncertain
