Gene
KWMTBOMO04242
Pre Gene Modal
BGIBMGA014260
Annotation
PREDICTED:_metabotropic_glutamate_receptor_2-like_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.769
Sequence
CDS
ATGTTACCCGGCTGCAAGTTTGCTTTACCGCTATTCATCATCATTCTGCAACATTCAGAGATGAAACACTTCGGATATAGTCAGCACTATGCTCCAACACTTGATGAGTCCGAATTTCACAGTAGAACATCGAGGCATCACCTTAGACACAAACATAAGCATAACCCAGAAATCTTGGACCAAACTTTTGTTGCTGACATACCAATATTTGCGAATCTAAAAGGAGTGGGTTTTATCGACAGAGAGTTCAATGAACCGGTATTTAAGAATGGTAAGGACTGGTTGAACATGTCTAGCGACCAGGTTGGAGAGATGAAGGACAAGAAACACCAACATTGGCCAATTAAAAGAGAAGCAGTTATTGAAGGAGATTTGGTACTGGGTGGTTTGATGATGGTGCACGAAAGATCTGATAATATGACCTGTGGTCCTGTGATGCCGCAAGGAGGAGTACAGGCTTTGGAAACTATGCTATACACGTTAGATATCATTAATAATAAGCAGGAGCTAATACCAGGCGTGGTGCTTGGAGCGCATATCTTAGATGACTGTGATAAGGACACGTATGGATTAGAAATGGCCGTAGACTTTATCAAAGGTTCAATAAGCAACATCGATGATGCAGAGTACGCTTGTAATGCGACCGCTGTGCGTAAAGTCATCTCAGGAGTGGTAGGTGCAGCATCCAGTGTGACGTCGGTGCAAGTCGCGAATCTGCTGAGACTGTTCAAAATACCGCAGGTTTCTTTCTTTTCCACATCACCGGAGCTGTCCAACAAAGCCCGTTTCGAGTACTTCACTAGGACAATACCTTCAGACCTTCACCAAGTGCGAGCGATGGTGGAAATCGTGAAGAAACTTGGCTGGAGCTACGTGTCTATTATTTACGAGGAGTCCAGTTACGGTATTAAGGCATTCGAAGAACTTGAAGCGCTTCTGGCTAGAAATGGCATCTGCATAGCTGTCAAAGAGAAGCTGGTAAAGGACTCTGGAGTTGCTGACGAGAAGGCATATGACACGATTGTCCAGAAATTGCTGACTAAGCCCCGGGCTAGAGGTGCAATAATCTTCGGTTCTGATCAAGAAGTGGCTGGTGTAATGAGAGCGGTGATACGTGGCAATGCCACAAGCTCCTTCAGCTGGGTGGGGTCTGACGGCTGGAGCGCGCGGTCTCTGGTGTCCGAAGGCAACGAGCAGGCAGTCGAAGGGACCATCAGCGTCCAGCCGCAAGCGAATGATGTGAGGGGCTTTAAAGAGTACTTTTTGAACCTGAATGTGAAGAACAATAAGAGGAACCCTTGGTTTGTTGAATTCTGGGAAGACCACTTCCAGTGCCGCTACCCAGGTAGTCCCAGAACTCCGTACAATGGGCAGTATTCCAAGACCTGCTCTGGACTTGAACGCCTTACCACCGAGAACACTGAGTTTGAGAGGCAATTGCAGTTCGTTTCTGATGCTGTGATGGCCTTCGCTTATGCTATACAGTAA
Protein
MLPGCKFALPLFIIILQHSEMKHFGYSQHYAPTLDESEFHSRTSRHHLRHKHKHNPEILDQTFVADIPIFANLKGVGFIDREFNEPVFKNGKDWLNMSSDQVGEMKDKKHQHWPIKREAVIEGDLVLGGLMMVHERSDNMTCGPVMPQGGVQALETMLYTLDIINNKQELIPGVVLGAHILDDCDKDTYGLEMAVDFIKGSISNIDDAEYACNATAVRKVISGVVGAASSVTSVQVANLLRLFKIPQVSFFSTSPELSNKARFEYFTRTIPSDLHQVRAMVEIVKKLGWSYVSIIYEESSYGIKAFEELEALLARNGICIAVKEKLVKDSGVADEKAYDTIVQKLLTKPRARGAIIFGSDQEVAGVMRAVIRGNATSSFSWVGSDGWSARSLVSEGNEQAVEGTISVQPQANDVRGFKEYFLNLNVKNNKRNPWFVEFWEDHFQCRYPGSPRTPYNGQYSKTCSGLERLTTENTEFERQLQFVSDAVMAFAYAIQ
Summary
Similarity
Belongs to the G-protein coupled receptor 3 family.
Uniprot
H9JXJ4
A0A2W1BFP1
A0A194RGF0
A0A194Q8J6
A0A212EHW5
A0A1A9VV31
+ More
A0A1I8M0J9 A0A195D7C2 A0A151WWG1 A0A1I8NMS5 A0A195DLK5 A0A195B3M6 A0A154PQC1 A0A3B0IZT4 A0A195FTU1 B4LLC1 B4GDF5 A0A2W1BGB3 B5DZA5 A0A0B4KF15 B4P2U6 A0A0K8U462 B7YZT0 A0A0P9ACD0 A0A0J9TWB7 A0A0R3NL13 A1Z7F5 B4J5X5 A0A0J9R8A8 A0A2A4JI84 Q70GQ8 B3N8N5 K7J0X5 A0A0R1DM99 B4HRY5 A0A1W4UJR8 B3MFX4 A1Z7F4 B4KP51 A0A088AQD5 A0A0Q5WKX7 Q75QW6 A0A1B0C9E3 A0A1J0FAU2 A0A1J0FAV9 A0A182MEY3 A0A182PRF3 A0A1W7R6Q5 T1I399 A0A0L7L9Z1 A0A1J1J939 W8BYX6 A0A084WKZ5 Q7Q9V1 A0A182YJ34 A0A182NLU4 A0A182R0R1 B4MQ18 A0A2H1VXT3 Q16UR9 A0A162NFS0 A0A0P5B4Y9 A0A0P5ZWN1 A0A0P5MEV6 A0A0P5NQX0 A0A0P5ZUU6 A0A0P6FQX3 A0A0N7ZY47 A0A0P5C1T2 A0A0P5BX88 A0A0P5PNH9 A0A0P6H331 A0A0P5XG85 A0A0P5WKR7 A0A0P5S503 A0A0P5AEQ7 A0A0P5MYY8 A0A0P4ZXW9 A0A0P5DZT9 A0A182X0Z4 A0A0P5VY62 A0A0P5AG80 A0A0P5W0C8 A0A0P5MY78 A0A0P5M842 A0A0P5MTW6 A0A0N8BX12 A0A0P5M855 A0A0P5M4G0 A0A0N7ZYK9 A0A0P5N2P5 A0A0N8AAD9 A0A0P5B456 A0A0P5P251 A0A0P5B5K5 A0A0P4ZXS8 A0A0P5PCA0 A0A0P5PG09 A0A0N8CRK3 A0A0N8CQ02
A0A1I8M0J9 A0A195D7C2 A0A151WWG1 A0A1I8NMS5 A0A195DLK5 A0A195B3M6 A0A154PQC1 A0A3B0IZT4 A0A195FTU1 B4LLC1 B4GDF5 A0A2W1BGB3 B5DZA5 A0A0B4KF15 B4P2U6 A0A0K8U462 B7YZT0 A0A0P9ACD0 A0A0J9TWB7 A0A0R3NL13 A1Z7F5 B4J5X5 A0A0J9R8A8 A0A2A4JI84 Q70GQ8 B3N8N5 K7J0X5 A0A0R1DM99 B4HRY5 A0A1W4UJR8 B3MFX4 A1Z7F4 B4KP51 A0A088AQD5 A0A0Q5WKX7 Q75QW6 A0A1B0C9E3 A0A1J0FAU2 A0A1J0FAV9 A0A182MEY3 A0A182PRF3 A0A1W7R6Q5 T1I399 A0A0L7L9Z1 A0A1J1J939 W8BYX6 A0A084WKZ5 Q7Q9V1 A0A182YJ34 A0A182NLU4 A0A182R0R1 B4MQ18 A0A2H1VXT3 Q16UR9 A0A162NFS0 A0A0P5B4Y9 A0A0P5ZWN1 A0A0P5MEV6 A0A0P5NQX0 A0A0P5ZUU6 A0A0P6FQX3 A0A0N7ZY47 A0A0P5C1T2 A0A0P5BX88 A0A0P5PNH9 A0A0P6H331 A0A0P5XG85 A0A0P5WKR7 A0A0P5S503 A0A0P5AEQ7 A0A0P5MYY8 A0A0P4ZXW9 A0A0P5DZT9 A0A182X0Z4 A0A0P5VY62 A0A0P5AG80 A0A0P5W0C8 A0A0P5MY78 A0A0P5M842 A0A0P5MTW6 A0A0N8BX12 A0A0P5M855 A0A0P5M4G0 A0A0N7ZYK9 A0A0P5N2P5 A0A0N8AAD9 A0A0P5B456 A0A0P5P251 A0A0P5B5K5 A0A0P4ZXS8 A0A0P5PCA0 A0A0P5PG09 A0A0N8CRK3 A0A0N8CQ02
Pubmed
EMBL
BABH01032726
BABH01032727
KZ150080
PZC73882.1
KQ460211
KPJ16524.1
+ More
KQ459302 KPJ01848.1 AGBW02014793 OWR41061.1 KQ976749 KYN08785.1 KQ982691 KYQ52223.1 KQ980765 KYN13374.1 KQ976641 KYM78799.1 KQ435036 KZC14082.1 OUUW01000001 SPP73687.1 KQ981272 KYN43846.1 CH940648 EDW60858.2 CH479181 EDW31631.1 KZ150487 PZC70703.1 CM000071 EDY68791.2 AE013599 AGB93339.1 CM000157 EDW89357.2 GDHF01030797 JAI21517.1 ACL83073.1 CH902619 KPU75778.1 CM002911 KMY92273.1 KMY92274.1 KMY92275.1 KRT01640.1 AAF59082.3 CH916367 EDW00818.1 KMY92276.1 NWSH01001473 PCG71140.1 AJ581534 CAE46392.1 CH954177 EDV59512.1 KRJ98364.1 CH480816 EDW46948.1 EDV35656.2 AAF59081.3 CH933808 EDW10117.2 KQS70875.1 AB161182 BAD08344.1 AJWK01002248 KU986887 APC26116.1 KU986890 APC26119.1 AXCM01007183 AXCM01007184 GEHC01000817 JAV46828.1 ACPB03010517 JTDY01002016 KOB72318.1 CVRI01000074 CRL08038.1 GAMC01002043 JAC04513.1 ATLV01024163 KE525350 KFB50889.1 AAAB01008900 EAA09380.4 AXCN02002332 CH963849 EDW74207.1 ODYU01005102 SOQ45645.1 CH477615 EAT38269.2 LRGB01000568 KZS17936.1 GDIP01189915 JAJ33487.1 GDIP01037990 JAM65725.1 GDIQ01159366 JAK92359.1 GDIQ01139066 JAL12660.1 GDIP01039190 JAM64525.1 GDIQ01044505 GDIQ01044504 JAN50233.1 GDIP01201161 JAJ22241.1 GDIP01194017 GDIP01192215 JAJ31187.1 GDIP01194016 GDIP01192214 JAJ29386.1 GDIQ01136566 JAL15160.1 GDIQ01046503 JAN48234.1 GDIP01084891 JAM18824.1 GDIP01084890 JAM18825.1 GDIQ01214878 GDIQ01109465 JAL42261.1 GDIP01199866 JAJ23536.1 GDIQ01157461 JAK94264.1 GDIP01206564 JAJ16838.1 GDIP01166826 JAJ56576.1 GDIP01093580 JAM10135.1 GDIP01204511 JAJ18891.1 GDIP01093578 JAM10137.1 GDIQ01160960 GDIQ01160959 JAK90766.1 GDIQ01159361 JAK92364.1 GDIQ01159362 JAK92363.1 GDIQ01136563 JAL15163.1 GDIQ01159360 JAK92365.1 GDIQ01160961 JAK90764.1 GDIP01199865 JAJ23537.1 GDIQ01159359 GDIQ01157464 JAK92366.1 GDIP01166825 JAJ56577.1 GDIP01189914 JAJ33488.1 GDIQ01139068 JAL12658.1 GDIP01189917 JAJ33485.1 GDIP01206563 JAJ16839.1 GDIQ01139069 GDIQ01136569 JAL15157.1 GDIQ01139071 JAL12655.1 GDIP01105823 JAL97891.1 GDIP01110231 GDIP01061384 JAL93483.1
KQ459302 KPJ01848.1 AGBW02014793 OWR41061.1 KQ976749 KYN08785.1 KQ982691 KYQ52223.1 KQ980765 KYN13374.1 KQ976641 KYM78799.1 KQ435036 KZC14082.1 OUUW01000001 SPP73687.1 KQ981272 KYN43846.1 CH940648 EDW60858.2 CH479181 EDW31631.1 KZ150487 PZC70703.1 CM000071 EDY68791.2 AE013599 AGB93339.1 CM000157 EDW89357.2 GDHF01030797 JAI21517.1 ACL83073.1 CH902619 KPU75778.1 CM002911 KMY92273.1 KMY92274.1 KMY92275.1 KRT01640.1 AAF59082.3 CH916367 EDW00818.1 KMY92276.1 NWSH01001473 PCG71140.1 AJ581534 CAE46392.1 CH954177 EDV59512.1 KRJ98364.1 CH480816 EDW46948.1 EDV35656.2 AAF59081.3 CH933808 EDW10117.2 KQS70875.1 AB161182 BAD08344.1 AJWK01002248 KU986887 APC26116.1 KU986890 APC26119.1 AXCM01007183 AXCM01007184 GEHC01000817 JAV46828.1 ACPB03010517 JTDY01002016 KOB72318.1 CVRI01000074 CRL08038.1 GAMC01002043 JAC04513.1 ATLV01024163 KE525350 KFB50889.1 AAAB01008900 EAA09380.4 AXCN02002332 CH963849 EDW74207.1 ODYU01005102 SOQ45645.1 CH477615 EAT38269.2 LRGB01000568 KZS17936.1 GDIP01189915 JAJ33487.1 GDIP01037990 JAM65725.1 GDIQ01159366 JAK92359.1 GDIQ01139066 JAL12660.1 GDIP01039190 JAM64525.1 GDIQ01044505 GDIQ01044504 JAN50233.1 GDIP01201161 JAJ22241.1 GDIP01194017 GDIP01192215 JAJ31187.1 GDIP01194016 GDIP01192214 JAJ29386.1 GDIQ01136566 JAL15160.1 GDIQ01046503 JAN48234.1 GDIP01084891 JAM18824.1 GDIP01084890 JAM18825.1 GDIQ01214878 GDIQ01109465 JAL42261.1 GDIP01199866 JAJ23536.1 GDIQ01157461 JAK94264.1 GDIP01206564 JAJ16838.1 GDIP01166826 JAJ56576.1 GDIP01093580 JAM10135.1 GDIP01204511 JAJ18891.1 GDIP01093578 JAM10137.1 GDIQ01160960 GDIQ01160959 JAK90766.1 GDIQ01159361 JAK92364.1 GDIQ01159362 JAK92363.1 GDIQ01136563 JAL15163.1 GDIQ01159360 JAK92365.1 GDIQ01160961 JAK90764.1 GDIP01199865 JAJ23537.1 GDIQ01159359 GDIQ01157464 JAK92366.1 GDIP01166825 JAJ56577.1 GDIP01189914 JAJ33488.1 GDIQ01139068 JAL12658.1 GDIP01189917 JAJ33485.1 GDIP01206563 JAJ16839.1 GDIQ01139069 GDIQ01136569 JAL15157.1 GDIQ01139071 JAL12655.1 GDIP01105823 JAL97891.1 GDIP01110231 GDIP01061384 JAL93483.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000078200
UP000095301
+ More
UP000078542 UP000075809 UP000095300 UP000078492 UP000078540 UP000076502 UP000268350 UP000078541 UP000008792 UP000008744 UP000001819 UP000000803 UP000002282 UP000007801 UP000001070 UP000218220 UP000008711 UP000002358 UP000001292 UP000192221 UP000009192 UP000005203 UP000092461 UP000075883 UP000075885 UP000015103 UP000037510 UP000183832 UP000030765 UP000007062 UP000076408 UP000075884 UP000075886 UP000007798 UP000008820 UP000076858 UP000076407
UP000078542 UP000075809 UP000095300 UP000078492 UP000078540 UP000076502 UP000268350 UP000078541 UP000008792 UP000008744 UP000001819 UP000000803 UP000002282 UP000007801 UP000001070 UP000218220 UP000008711 UP000002358 UP000001292 UP000192221 UP000009192 UP000005203 UP000092461 UP000075883 UP000075885 UP000015103 UP000037510 UP000183832 UP000030765 UP000007062 UP000076408 UP000075884 UP000075886 UP000007798 UP000008820 UP000076858 UP000076407
Interpro
SUPFAM
SSF53822
SSF53822
Gene 3D
ProteinModelPortal
H9JXJ4
A0A2W1BFP1
A0A194RGF0
A0A194Q8J6
A0A212EHW5
A0A1A9VV31
+ More
A0A1I8M0J9 A0A195D7C2 A0A151WWG1 A0A1I8NMS5 A0A195DLK5 A0A195B3M6 A0A154PQC1 A0A3B0IZT4 A0A195FTU1 B4LLC1 B4GDF5 A0A2W1BGB3 B5DZA5 A0A0B4KF15 B4P2U6 A0A0K8U462 B7YZT0 A0A0P9ACD0 A0A0J9TWB7 A0A0R3NL13 A1Z7F5 B4J5X5 A0A0J9R8A8 A0A2A4JI84 Q70GQ8 B3N8N5 K7J0X5 A0A0R1DM99 B4HRY5 A0A1W4UJR8 B3MFX4 A1Z7F4 B4KP51 A0A088AQD5 A0A0Q5WKX7 Q75QW6 A0A1B0C9E3 A0A1J0FAU2 A0A1J0FAV9 A0A182MEY3 A0A182PRF3 A0A1W7R6Q5 T1I399 A0A0L7L9Z1 A0A1J1J939 W8BYX6 A0A084WKZ5 Q7Q9V1 A0A182YJ34 A0A182NLU4 A0A182R0R1 B4MQ18 A0A2H1VXT3 Q16UR9 A0A162NFS0 A0A0P5B4Y9 A0A0P5ZWN1 A0A0P5MEV6 A0A0P5NQX0 A0A0P5ZUU6 A0A0P6FQX3 A0A0N7ZY47 A0A0P5C1T2 A0A0P5BX88 A0A0P5PNH9 A0A0P6H331 A0A0P5XG85 A0A0P5WKR7 A0A0P5S503 A0A0P5AEQ7 A0A0P5MYY8 A0A0P4ZXW9 A0A0P5DZT9 A0A182X0Z4 A0A0P5VY62 A0A0P5AG80 A0A0P5W0C8 A0A0P5MY78 A0A0P5M842 A0A0P5MTW6 A0A0N8BX12 A0A0P5M855 A0A0P5M4G0 A0A0N7ZYK9 A0A0P5N2P5 A0A0N8AAD9 A0A0P5B456 A0A0P5P251 A0A0P5B5K5 A0A0P4ZXS8 A0A0P5PCA0 A0A0P5PG09 A0A0N8CRK3 A0A0N8CQ02
A0A1I8M0J9 A0A195D7C2 A0A151WWG1 A0A1I8NMS5 A0A195DLK5 A0A195B3M6 A0A154PQC1 A0A3B0IZT4 A0A195FTU1 B4LLC1 B4GDF5 A0A2W1BGB3 B5DZA5 A0A0B4KF15 B4P2U6 A0A0K8U462 B7YZT0 A0A0P9ACD0 A0A0J9TWB7 A0A0R3NL13 A1Z7F5 B4J5X5 A0A0J9R8A8 A0A2A4JI84 Q70GQ8 B3N8N5 K7J0X5 A0A0R1DM99 B4HRY5 A0A1W4UJR8 B3MFX4 A1Z7F4 B4KP51 A0A088AQD5 A0A0Q5WKX7 Q75QW6 A0A1B0C9E3 A0A1J0FAU2 A0A1J0FAV9 A0A182MEY3 A0A182PRF3 A0A1W7R6Q5 T1I399 A0A0L7L9Z1 A0A1J1J939 W8BYX6 A0A084WKZ5 Q7Q9V1 A0A182YJ34 A0A182NLU4 A0A182R0R1 B4MQ18 A0A2H1VXT3 Q16UR9 A0A162NFS0 A0A0P5B4Y9 A0A0P5ZWN1 A0A0P5MEV6 A0A0P5NQX0 A0A0P5ZUU6 A0A0P6FQX3 A0A0N7ZY47 A0A0P5C1T2 A0A0P5BX88 A0A0P5PNH9 A0A0P6H331 A0A0P5XG85 A0A0P5WKR7 A0A0P5S503 A0A0P5AEQ7 A0A0P5MYY8 A0A0P4ZXW9 A0A0P5DZT9 A0A182X0Z4 A0A0P5VY62 A0A0P5AG80 A0A0P5W0C8 A0A0P5MY78 A0A0P5M842 A0A0P5MTW6 A0A0N8BX12 A0A0P5M855 A0A0P5M4G0 A0A0N7ZYK9 A0A0P5N2P5 A0A0N8AAD9 A0A0P5B456 A0A0P5P251 A0A0P5B5K5 A0A0P4ZXS8 A0A0P5PCA0 A0A0P5PG09 A0A0N8CRK3 A0A0N8CQ02
PDB
2E4Y
E-value=3.87649e-73,
Score=700
Ontologies
GO
Topology
Subcellular location
Cell membrane
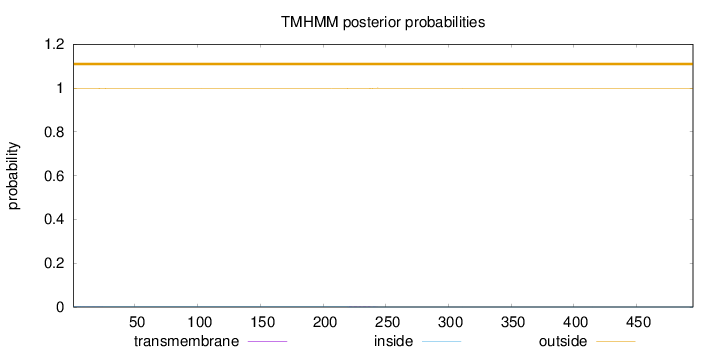
Length:
495
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0421900000000001
Exp number, first 60 AAs:
0.0122
Total prob of N-in:
0.00215
outside
1 - 495
Population Genetic Test Statistics
Pi
251.295779
Theta
158.657919
Tajima's D
1.338229
CLR
0.309405
CSRT
0.749712514374281
Interpretation
Uncertain
