Gene
KWMTBOMO04241
Annotation
reverse_transcriptase_[Bombyx_mori]
Full name
Probable RNA-directed DNA polymerase from transposon BS
Alternative Name
Reverse transcriptase
Location in the cell
Mitochondrial Reliability : 2.13
Sequence
CDS
ATGGAAGTTAAAGACTTGATCAAAGACCTACGTCCTCGCAAGGCTCCCGGTTCCGACGGTATATCTAACCGCGTTATTAAACTTCTACCCGTCCAACTCATCGTGATGTTGGCATCTATTTTCAATGCCGCTATGGCGAACTGTATCTTTCCCGCGGTGTGGAAAGAAGCGGACGTTATCGGCATACATAAACCCGGTAAACCAAAAAATCATCCGACGAGCTACCGCCCGATTAGCCTCCTCATGTCTCTAGGCAAACTGTATGAGCGTCAGCTCTACAAACGCCTCAGAGACTTCGTCTCATCCAAGGGCATTCTCATCGATGAACAATTCGGATTCCGTACAAATCACTCATGCGTTCAACAGGTGCACCGCCTCACGGAGCACATTCTTGTGGGGCTTAATCGACCAAAACCGTTATACACGGGAGCTATCTTCTTCGACGTCGCAAAAGCGTTCGATAAAGTCTGGCACAATGGTTTGATTTTCAAACTATTCAACATGGGCGTGCCGGATAGTCTCGTGCTCATCATACGGGACTTCTTGTCGAACCGCTCTTTTCGATATCGAGTCGAGGGAACCCGCTCCTCCCCACGACCTCTCACAGCTGGAGTCCCGCAAGGCTCTGTCCTCTCACCCCTCCTATTTAGCTTATTCGTCAACGATGTTCCCCGGTCGCCGCCGACCCATTTAGCTTTATTCGCCGACGACACGACTGTTTACTATTCTAGTAGAAATAAGTCCCTAATCGCGAAGAAGCTTCAGAGCGCAGCCCTAGCCCTAGGACAGTGGTTCCGAAAATGGCGCATAGACATCAACCCAGCGAAAAGTACTGCGGTGCTCTTTCAGAGGGGAAGCTCCACACGGATTTCCTCCCGGATTAGGAGGAGGAATCTCACACCCCCGATTACTCTCTTTAGACAACCCATACCCTGGGCCAGGAAGGTCAAGTACCTGGGCGTTACCCTGGATGCATCGATGACATTCCGCCCGCATATAAAATCAGTCCGTGACCGTGCCGCGTTTATTCTCGGTAGACTCTTCCCCATGATCTGTAAGCGGAGTAAAATGTCCCTTCGGAACAAGGTGACACTTTACAAAACTTGCATAAGGCCCGTCATGACTTACGCGAGTGTGGTGTTCGCTCACGCGGCCCGCACACACATAGACACCCTCCAATCCCTACAATCCCGCTTTTGCAGGTTAGCTGTCGGGGCTCCGTGGTTCGTGAGGAACGTTGACCTACACGACGACCTGGGCCTCGAATCAATTCGGAAATACATGAAGTCAGCGTCGGAACGATACTTCGATAAGGCTATGCGTCATGATAATCGCCTTATCGTTGCCGCCGCTGACTACTCCCCGAATCCTGATCATGCAGGAGCCAGTCACCGTCGACGCCCTAGACACGTCCTTACGGATCCATCAGATCCAATAACCTTTGCATTAGATGCCTTCAGCTCTAATACTAGGGGCAGGCTTAGGGACCCCGGTAACCGTACTCGTCGAACTCGACAAAGAGGTCGACGTGCAACCTAA
Protein
MEVKDLIKDLRPRKAPGSDGISNRVIKLLPVQLIVMLASIFNAAMANCIFPAVWKEADVIGIHKPGKPKNHPTSYRPISLLMSLGKLYERQLYKRLRDFVSSKGILIDEQFGFRTNHSCVQQVHRLTEHILVGLNRPKPLYTGAIFFDVAKAFDKVWHNGLIFKLFNMGVPDSLVLIIRDFLSNRSFRYRVEGTRSSPRPLTAGVPQGSVLSPLLFSLFVNDVPRSPPTHLALFADDTTVYYSSRNKSLIAKKLQSAALALGQWFRKWRIDINPAKSTAVLFQRGSSTRISSRIRRRNLTPPITLFRQPIPWARKVKYLGVTLDASMTFRPHIKSVRDRAAFILGRLFPMICKRSKMSLRNKVTLYKTCIRPVMTYASVVFAHAARTHIDTLQSLQSRFCRLAVGAPWFVRNVDLHDDLGLESIRKYMKSASERYFDKAMRHDNRLIVAAADYSPNPDHAGASHRRRPRHVLTDPSDPITFALDAFSSNTRGRLRDPGNRTRRTRQRGRRAT
Summary
Catalytic Activity
a 2'-deoxyribonucleoside 5'-triphosphate + DNA(n) = diphosphate + DNA(n+1)
Cofactor
Mg(2+)
Mn(2+)
Mn(2+)
Keywords
Nucleotidyltransferase
RNA-directed DNA polymerase
Transferase
Transposable element
Feature
chain Probable RNA-directed DNA polymerase from transposon BS
Uniprot
Q93137
Q9XXW0
Q9BPQ0
K9N5J5
A0A0N0PA09
A0A0N1PFV1
+ More
O96546 O96544 A0A194Q1H8 A0A1Y1NA23 V5GNF3 A0A2S2PR14 V5GNJ2 A0A139WA11 X1WXU3 X1WLB2 X1WMU5 A0A139W8G9 A0A2S2PX53 J9KKL3 A0A2S2QTZ8 X1WWC4 A0A2S2PH29 A0A2S2Q823 A0A1Y1M7E9 A0A2S2QJM7 J9KC83 A0A087U2S8 A0A2S2PPK8 O76197 A0A023EYS8 A0A2S2NGV8 A0A224XF24 A0A2S2Q352 A0A2S2N814 A0A2S2QEE0 A0A023EZE6 V5I850 A0A2J7RL12 A0A2J7R7D0 A0A1Y1LKN6 A0A2J7RS81 A0A2S2RAN7 A0A2J7PVX3 J9LC15 A0A2H8TWD9 A0A2J7Q627 J9M522 A0A2H8TES1 A0A2S2NJB5 A0A2J7R8R8 A0A2H8U0H5 J9KCZ4 A0A0A9YJ00 A0A2A4K8E7 A0A2M4BDV6 A0A2J7PDM0 A0A2M4BD67 A0A023F6S4 X1WZH0 A0A0C9RJA4 A0A2J7QPI2 A0A087V0P8 X1WN18 A0A2J7R579 A0A2J7PQB5 A0A2J7RIW7 A0A2J7R4B1 A0A0A9YU04 A0A2J7R4B7 A0A3N5Z7I3 A0A2J7PEB1 A0A2J7RM10 A0A2J7PMP6 A0A2J7Q4F7 X1WNI2 A0A2S2PBB4 A0A069DZC5 A0A2S2R9D4 A0A0J7KBP6 A0A2J7Q059 A0A2J7QB68 X1WTA1 A0A2J7QUJ9 A0A2P8Z520 A0A087T1W0 A0A0J7K4E0 A0A069DW01 A0A2J7RB54 A0A2J7QTW2 A0A2S2PYN7 A0A2J7PMP9 A0A2A4K918 A0A2J7RPU2 A0A2J7PU96 A0A1Y1KFR3 A0A2S2P0Y0 A0A2J7RS77 J9LC21 Q95SX7
O96546 O96544 A0A194Q1H8 A0A1Y1NA23 V5GNF3 A0A2S2PR14 V5GNJ2 A0A139WA11 X1WXU3 X1WLB2 X1WMU5 A0A139W8G9 A0A2S2PX53 J9KKL3 A0A2S2QTZ8 X1WWC4 A0A2S2PH29 A0A2S2Q823 A0A1Y1M7E9 A0A2S2QJM7 J9KC83 A0A087U2S8 A0A2S2PPK8 O76197 A0A023EYS8 A0A2S2NGV8 A0A224XF24 A0A2S2Q352 A0A2S2N814 A0A2S2QEE0 A0A023EZE6 V5I850 A0A2J7RL12 A0A2J7R7D0 A0A1Y1LKN6 A0A2J7RS81 A0A2S2RAN7 A0A2J7PVX3 J9LC15 A0A2H8TWD9 A0A2J7Q627 J9M522 A0A2H8TES1 A0A2S2NJB5 A0A2J7R8R8 A0A2H8U0H5 J9KCZ4 A0A0A9YJ00 A0A2A4K8E7 A0A2M4BDV6 A0A2J7PDM0 A0A2M4BD67 A0A023F6S4 X1WZH0 A0A0C9RJA4 A0A2J7QPI2 A0A087V0P8 X1WN18 A0A2J7R579 A0A2J7PQB5 A0A2J7RIW7 A0A2J7R4B1 A0A0A9YU04 A0A2J7R4B7 A0A3N5Z7I3 A0A2J7PEB1 A0A2J7RM10 A0A2J7PMP6 A0A2J7Q4F7 X1WNI2 A0A2S2PBB4 A0A069DZC5 A0A2S2R9D4 A0A0J7KBP6 A0A2J7Q059 A0A2J7QB68 X1WTA1 A0A2J7QUJ9 A0A2P8Z520 A0A087T1W0 A0A0J7K4E0 A0A069DW01 A0A2J7RB54 A0A2J7QTW2 A0A2S2PYN7 A0A2J7PMP9 A0A2A4K918 A0A2J7RPU2 A0A2J7PU96 A0A1Y1KFR3 A0A2S2P0Y0 A0A2J7RS77 J9LC21 Q95SX7
EC Number
2.7.7.49
Pubmed
EMBL
U07847
AAA17752.1
AB018558
BAA76304.1
AB055391
BAB21761.1
+ More
KC165845 AFY12622.1 KQ459259 KPJ02068.1 KQ459265 KPJ02064.1 AF081103 AAC72921.1 AF081101 AAC72919.1 KQ459580 KPI99178.1 GEZM01008457 JAV94772.1 GALX01005359 JAB63107.1 GGMR01019198 MBY31817.1 GALX01005299 JAB63167.1 KQ971729 KYB24751.1 ABLF02008933 ABLF02042518 ABLF02057746 ABLF02014862 ABLF02014866 ABLF02061908 KQ973342 KXZ75569.1 GGMS01000737 MBY69940.1 ABLF02029306 ABLF02029314 ABLF02060138 GGMS01012005 MBY81208.1 ABLF02003961 ABLF02059872 GGMR01016114 MBY28733.1 GGMS01004685 MBY73888.1 GEZM01038405 JAV81653.1 GGMS01008766 MBY77969.1 ABLF02033757 KK117880 KFM71667.1 GGMR01018716 MBY31335.1 AF012049 AAC24972.1 GBBI01004395 JAC14317.1 GGMR01003407 MBY16026.1 GFTR01008038 JAW08388.1 GGMS01002955 MBY72158.1 GGMR01000676 MBY13295.1 GGMS01006359 MBY75562.1 GBBI01004571 JAC14141.1 GALX01005300 JAB63166.1 NEVH01002704 PNF41489.1 NEVH01006736 PNF36716.1 GEZM01053022 JAV74219.1 NEVH01000269 PNF43694.1 GGMS01017914 MBY87117.1 NEVH01020940 PNF20481.1 ABLF02020655 ABLF02039168 ABLF02061060 GFXV01006424 MBW18229.1 NEVH01017542 PNF24034.1 ABLF02024246 GFXV01000774 MBW12579.1 GGMR01004661 MBY17280.1 NEVH01006721 PNF37225.1 GFXV01007093 MBW18898.1 ABLF02008463 GBHO01010547 GBHO01010546 JAG33057.1 JAG33058.1 NWSH01000032 PCG80497.1 GGFJ01001857 MBW50998.1 NEVH01026386 PNF14434.1 GGFJ01001858 MBW50999.1 GBBI01001646 JAC17066.1 ABLF02034467 GBYB01008290 JAG78057.1 NEVH01012089 PNF30491.1 KL813758 KFM83187.1 ABLF02021177 ABLF02027367 ABLF02066338 NEVH01007393 PNF35987.1 NEVH01022639 PNF18527.1 NEVH01003017 PNF40781.1 NEVH01007578 PNF35646.1 GBHO01021814 GBHO01010549 JAG21790.1 JAG33055.1 NEVH01007443 PNF35684.1 RPOV01000134 RPJ78669.1 NEVH01026106 PNF14658.1 NEVH01021586 NEVH01006600 NEVH01002584 PNF19589.1 PNF37419.1 PNF41868.1 NEVH01023962 PNF17612.1 NEVH01018383 PNF23463.1 ABLF02008153 ABLF02008462 ABLF02008464 GGMR01014053 MBY26672.1 GBGD01000880 JAC88009.1 GGMS01017365 MBY86568.1 LBMM01010106 KMQ87671.1 NEVH01020071 PNF21969.1 NEVH01016302 PNF25823.1 ABLF02037660 NEVH01010546 PNF32261.1 PYGN01000192 PSN51582.1 KK113014 KFM59099.1 LBMM01014395 KMQ85192.1 GBGD01000799 JAC88090.1 NEVH01005938 PNF38077.1 NEVH01011193 PNF32021.1 GGMS01001277 MBY70480.1 NEVH01023961 PNF17613.1 PCG80496.1 NEVH01001355 PNF42855.1 NEVH01021205 PNF19912.1 GEZM01084969 JAV60363.1 GGMR01010373 MBY22992.1 NEVH01000280 PNF43672.1 ABLF02036316 ABLF02036319 ABLF02048663 ABLF02066977 AY060438 AAL25477.1
KC165845 AFY12622.1 KQ459259 KPJ02068.1 KQ459265 KPJ02064.1 AF081103 AAC72921.1 AF081101 AAC72919.1 KQ459580 KPI99178.1 GEZM01008457 JAV94772.1 GALX01005359 JAB63107.1 GGMR01019198 MBY31817.1 GALX01005299 JAB63167.1 KQ971729 KYB24751.1 ABLF02008933 ABLF02042518 ABLF02057746 ABLF02014862 ABLF02014866 ABLF02061908 KQ973342 KXZ75569.1 GGMS01000737 MBY69940.1 ABLF02029306 ABLF02029314 ABLF02060138 GGMS01012005 MBY81208.1 ABLF02003961 ABLF02059872 GGMR01016114 MBY28733.1 GGMS01004685 MBY73888.1 GEZM01038405 JAV81653.1 GGMS01008766 MBY77969.1 ABLF02033757 KK117880 KFM71667.1 GGMR01018716 MBY31335.1 AF012049 AAC24972.1 GBBI01004395 JAC14317.1 GGMR01003407 MBY16026.1 GFTR01008038 JAW08388.1 GGMS01002955 MBY72158.1 GGMR01000676 MBY13295.1 GGMS01006359 MBY75562.1 GBBI01004571 JAC14141.1 GALX01005300 JAB63166.1 NEVH01002704 PNF41489.1 NEVH01006736 PNF36716.1 GEZM01053022 JAV74219.1 NEVH01000269 PNF43694.1 GGMS01017914 MBY87117.1 NEVH01020940 PNF20481.1 ABLF02020655 ABLF02039168 ABLF02061060 GFXV01006424 MBW18229.1 NEVH01017542 PNF24034.1 ABLF02024246 GFXV01000774 MBW12579.1 GGMR01004661 MBY17280.1 NEVH01006721 PNF37225.1 GFXV01007093 MBW18898.1 ABLF02008463 GBHO01010547 GBHO01010546 JAG33057.1 JAG33058.1 NWSH01000032 PCG80497.1 GGFJ01001857 MBW50998.1 NEVH01026386 PNF14434.1 GGFJ01001858 MBW50999.1 GBBI01001646 JAC17066.1 ABLF02034467 GBYB01008290 JAG78057.1 NEVH01012089 PNF30491.1 KL813758 KFM83187.1 ABLF02021177 ABLF02027367 ABLF02066338 NEVH01007393 PNF35987.1 NEVH01022639 PNF18527.1 NEVH01003017 PNF40781.1 NEVH01007578 PNF35646.1 GBHO01021814 GBHO01010549 JAG21790.1 JAG33055.1 NEVH01007443 PNF35684.1 RPOV01000134 RPJ78669.1 NEVH01026106 PNF14658.1 NEVH01021586 NEVH01006600 NEVH01002584 PNF19589.1 PNF37419.1 PNF41868.1 NEVH01023962 PNF17612.1 NEVH01018383 PNF23463.1 ABLF02008153 ABLF02008462 ABLF02008464 GGMR01014053 MBY26672.1 GBGD01000880 JAC88009.1 GGMS01017365 MBY86568.1 LBMM01010106 KMQ87671.1 NEVH01020071 PNF21969.1 NEVH01016302 PNF25823.1 ABLF02037660 NEVH01010546 PNF32261.1 PYGN01000192 PSN51582.1 KK113014 KFM59099.1 LBMM01014395 KMQ85192.1 GBGD01000799 JAC88090.1 NEVH01005938 PNF38077.1 NEVH01011193 PNF32021.1 GGMS01001277 MBY70480.1 NEVH01023961 PNF17613.1 PCG80496.1 NEVH01001355 PNF42855.1 NEVH01021205 PNF19912.1 GEZM01084969 JAV60363.1 GGMR01010373 MBY22992.1 NEVH01000280 PNF43672.1 ABLF02036316 ABLF02036319 ABLF02048663 ABLF02066977 AY060438 AAL25477.1
Proteomes
Pfam
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
Q93137
Q9XXW0
Q9BPQ0
K9N5J5
A0A0N0PA09
A0A0N1PFV1
+ More
O96546 O96544 A0A194Q1H8 A0A1Y1NA23 V5GNF3 A0A2S2PR14 V5GNJ2 A0A139WA11 X1WXU3 X1WLB2 X1WMU5 A0A139W8G9 A0A2S2PX53 J9KKL3 A0A2S2QTZ8 X1WWC4 A0A2S2PH29 A0A2S2Q823 A0A1Y1M7E9 A0A2S2QJM7 J9KC83 A0A087U2S8 A0A2S2PPK8 O76197 A0A023EYS8 A0A2S2NGV8 A0A224XF24 A0A2S2Q352 A0A2S2N814 A0A2S2QEE0 A0A023EZE6 V5I850 A0A2J7RL12 A0A2J7R7D0 A0A1Y1LKN6 A0A2J7RS81 A0A2S2RAN7 A0A2J7PVX3 J9LC15 A0A2H8TWD9 A0A2J7Q627 J9M522 A0A2H8TES1 A0A2S2NJB5 A0A2J7R8R8 A0A2H8U0H5 J9KCZ4 A0A0A9YJ00 A0A2A4K8E7 A0A2M4BDV6 A0A2J7PDM0 A0A2M4BD67 A0A023F6S4 X1WZH0 A0A0C9RJA4 A0A2J7QPI2 A0A087V0P8 X1WN18 A0A2J7R579 A0A2J7PQB5 A0A2J7RIW7 A0A2J7R4B1 A0A0A9YU04 A0A2J7R4B7 A0A3N5Z7I3 A0A2J7PEB1 A0A2J7RM10 A0A2J7PMP6 A0A2J7Q4F7 X1WNI2 A0A2S2PBB4 A0A069DZC5 A0A2S2R9D4 A0A0J7KBP6 A0A2J7Q059 A0A2J7QB68 X1WTA1 A0A2J7QUJ9 A0A2P8Z520 A0A087T1W0 A0A0J7K4E0 A0A069DW01 A0A2J7RB54 A0A2J7QTW2 A0A2S2PYN7 A0A2J7PMP9 A0A2A4K918 A0A2J7RPU2 A0A2J7PU96 A0A1Y1KFR3 A0A2S2P0Y0 A0A2J7RS77 J9LC21 Q95SX7
O96546 O96544 A0A194Q1H8 A0A1Y1NA23 V5GNF3 A0A2S2PR14 V5GNJ2 A0A139WA11 X1WXU3 X1WLB2 X1WMU5 A0A139W8G9 A0A2S2PX53 J9KKL3 A0A2S2QTZ8 X1WWC4 A0A2S2PH29 A0A2S2Q823 A0A1Y1M7E9 A0A2S2QJM7 J9KC83 A0A087U2S8 A0A2S2PPK8 O76197 A0A023EYS8 A0A2S2NGV8 A0A224XF24 A0A2S2Q352 A0A2S2N814 A0A2S2QEE0 A0A023EZE6 V5I850 A0A2J7RL12 A0A2J7R7D0 A0A1Y1LKN6 A0A2J7RS81 A0A2S2RAN7 A0A2J7PVX3 J9LC15 A0A2H8TWD9 A0A2J7Q627 J9M522 A0A2H8TES1 A0A2S2NJB5 A0A2J7R8R8 A0A2H8U0H5 J9KCZ4 A0A0A9YJ00 A0A2A4K8E7 A0A2M4BDV6 A0A2J7PDM0 A0A2M4BD67 A0A023F6S4 X1WZH0 A0A0C9RJA4 A0A2J7QPI2 A0A087V0P8 X1WN18 A0A2J7R579 A0A2J7PQB5 A0A2J7RIW7 A0A2J7R4B1 A0A0A9YU04 A0A2J7R4B7 A0A3N5Z7I3 A0A2J7PEB1 A0A2J7RM10 A0A2J7PMP6 A0A2J7Q4F7 X1WNI2 A0A2S2PBB4 A0A069DZC5 A0A2S2R9D4 A0A0J7KBP6 A0A2J7Q059 A0A2J7QB68 X1WTA1 A0A2J7QUJ9 A0A2P8Z520 A0A087T1W0 A0A0J7K4E0 A0A069DW01 A0A2J7RB54 A0A2J7QTW2 A0A2S2PYN7 A0A2J7PMP9 A0A2A4K918 A0A2J7RPU2 A0A2J7PU96 A0A1Y1KFR3 A0A2S2P0Y0 A0A2J7RS77 J9LC21 Q95SX7
Ontologies
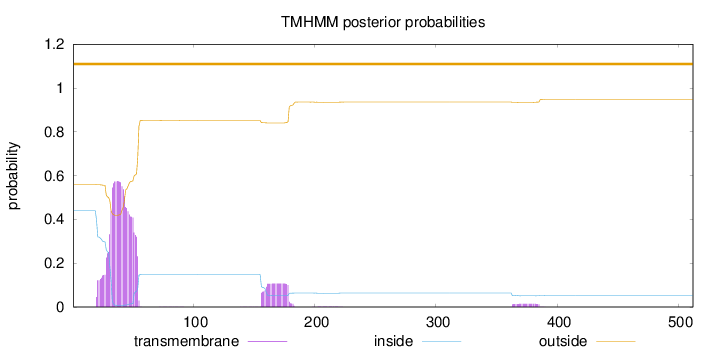
Topology
Length:
512
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
15.60247
Exp number, first 60 AAs:
12.86793
Total prob of N-in:
0.43959
POSSIBLE N-term signal
sequence
outside
1 - 512
Population Genetic Test Statistics
Pi
45.260368
Theta
32.40162
Tajima's D
2.959471
CLR
0.004462
CSRT
0.976951152442378
Interpretation
Uncertain
