Gene
KWMTBOMO04237
Pre Gene Modal
BGIBMGA014259
Annotation
PREDICTED:_inorganic_phosphate_transporter_1_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.978
Sequence
CDS
ATGCCAGGAGAGAAGGCTGTCTACAAGGGGGTACCAGCGGATGACAAATATGAAGGGTCATCACCAAAAATTCCGGCACCAAAAGGCTTTGGCACCCGCCACACTCAAGCTTTGGTACTGTTTATATGTTTAACCGTGGCGTACACCATGAGGGCTCAGCTCAGCGTGTCTATGGTAGCCATAACTGGTCACGCGGTCACAGAAAGTGAACTCTGCAGTAATGTAAGCACCAGTGAGAATACCAGCTGTGTTACAGAAGAAATCAGCGCCTGGAGTGTTTATCGTACGTACAACTGGAATAAATCGACTCAAGAAATGATATTGTTCGCGTTCTTCGTCGGCTACACCACCATGATGATCCCGATGGGTATAATAGCGCAGAAATTCGGTGGAAAATTTCCGATCAGTGTGGCGCTATTCGTTAATGGAGTTGTCTCTATCATCAGCCCTTGGATCCCTTTATTTAGCGGATGGATGGGTCTGGCCACATGTCGATTGATTCAAGGGATGACACAAGCAGCTCTCTATCCGAGCATGCACACAATGCTGGGTAAATGGGCACCGTTGAGTGAAAGAGGAAGATTGAGCACTTATGTCTACACGGGATCTCAATTCGGAACAATCTTGATCTTCGAAATATCAGGACTATTTGCTGGTACACCGGTCCTTGGTTGGCCCGCTATATTTTGGTTATGCGGTATCCTCAGTTTTGCCAGTTTCGGTCTTCTTATCTGGTTCGTCGCTCCGTCTCCTCGCGAACATCCGACCATAAGTGCTGAAGAATTATCTTACATTACGCAAGATGGAAATGTTGATGCTTTCCCGAAGAAACGCAGAACGCCATGGACGCATATCCTCACTAGCACAGCAGTTTGGGGTCTGGTAGTGAGCCACGCCGGGAGTTCGGCAGGATACCTTCTAGTCCTCACTCAGATGCCAACGTACATGAACGAAATACTTAACGTAAACATTGAAACTAATGGAATCTACTCATCCCTTCCTTACTTATCGATGTTCACGATGGCTATATTTTTCGGCTACATATCTGATTACTTACTGAACAAAAAGATTATGACAGTAGCCAACATCAGAAGAACAGCGAATACCATAGGAATGGTACTATCTGGGTTTTTCTTGATTGGATTCTGCTACGTGGATAAGGCCTGGTTGGCTGTATTGCTGCTAATGATATCATTTGGCATGCATTCTGCTGTTCACACTGGATTCCATATAAATCAAATAGACTTGTCGCCGAACTTTGCTGGTCCCATCATGTCTCTGGGAAACATGGTGGCGAACCTGACCAGTCTGCTGGTACCGGTCATTGTTTCAAACATCGTGAAAGATGATTTGAAAAATCCGAGCAGATGGCAAATAGTGTTCCTCATAACCGTGTCCTTCCAGTTCATAACCAATGCTGTCTTCGTGATATTCGTGAAAGGGTCGGTGCAGCCATGGAACTTTCACGACGAAGAAAGAATCGGTACAGGTGTAGAAGAATTAAAATCGCTTAACGACCACAAAAACAGCATCAAAGAAAAGGAGAAATTAGGAAATTCCTGA
Protein
MPGEKAVYKGVPADDKYEGSSPKIPAPKGFGTRHTQALVLFICLTVAYTMRAQLSVSMVAITGHAVTESELCSNVSTSENTSCVTEEISAWSVYRTYNWNKSTQEMILFAFFVGYTTMMIPMGIIAQKFGGKFPISVALFVNGVVSIISPWIPLFSGWMGLATCRLIQGMTQAALYPSMHTMLGKWAPLSERGRLSTYVYTGSQFGTILIFEISGLFAGTPVLGWPAIFWLCGILSFASFGLLIWFVAPSPREHPTISAEELSYITQDGNVDAFPKKRRTPWTHILTSTAVWGLVVSHAGSSAGYLLVLTQMPTYMNEILNVNIETNGIYSSLPYLSMFTMAIFFGYISDYLLNKKIMTVANIRRTANTIGMVLSGFFLIGFCYVDKAWLAVLLLMISFGMHSAVHTGFHINQIDLSPNFAGPIMSLGNMVANLTSLLVPVIVSNIVKDDLKNPSRWQIVFLITVSFQFITNAVFVIFVKGSVQPWNFHDEERIGTGVEELKSLNDHKNSIKEKEKLGNS
Summary
Uniprot
H9JXJ3
A0A2W1BM66
A0A2A4JR34
A0A2H1W147
A0A2H1V191
A0A0N0PCS9
+ More
A0A194Q007 A0A3S2LCW7 A0A2W1BHP9 A0A2W1BGX7 A0A194PZ61 A0A0N1IEP7 A0A2H1X2V7 A0A2H1VNZ8 A0A2A4JIZ6 A0A2H1VNW8 A0A2H1VP06 B0WB17 A0A2H1VPY0 A0A194Q004 A0A1Q3FPC2 A0A336LY62 A0A2H1VP00 A0A336LZS5 A0A2A4JKI4 A0A067QS64 A0A0N8P0I5 B3MI17 A0A1Q3FPF7 A0A1L8DE98 A0A1W4UP57 A0A2J7Q377 A0A2A4JKL3 Q17LW3 A0A1J1IVE3 A0A2J7Q364 A0A2M4CLX1 A0A0K8VIK3 A0A182LMR0 Q5BIE4 A0A0K8UFD6 A0A182Q5D9 A0A1S4GZN6 Q7JRA7 A0A182XNS8 A0A182U715 A0A2M4AF33 A0A0J9RHI1 B4NSL6 A0A034V4D0 A0A182VKS3 A0A182FN64 A0A194RFB6 W8AL50 W8AX47 A0A182Y0D9 W5JTB8 A0A182GLJ7 A0A2A4JRS5 A0A2M4AFD6 B4P485 A0A182I7D3 A0A194RFZ7 B3NKB8 A0A182K1W8 A0A0R1DS37 A0A0Q5VM65 B4HP21 A0A194PYB2 A0A182RI61 A0A212FKV3 A0A182PDV6 A0A0A1XJ44 A0A182MZ46 A0A0A1XHS7 A0A182W6C0 Q292H7 A0A2M3ZEY8 A0A2M4BLU2 T1PF36 A0A3B0JJR4 A0A1A9VA36 B4GDI7 A0A034VUA9 A0A1A9XAR9 A0A0R3NQT8 A0A1B0BN67 B4P486 A0A2A4JIL8 A0A2M4BMD9 A0A0L7R1A4 A0A151XIZ3 B3NKB7 A0A1A9WYG2 A0A2M4BMA3 A0A0A1XMC1 A0A182JDX2 A0A212FBF2 A0A0J9RG84
A0A194Q007 A0A3S2LCW7 A0A2W1BHP9 A0A2W1BGX7 A0A194PZ61 A0A0N1IEP7 A0A2H1X2V7 A0A2H1VNZ8 A0A2A4JIZ6 A0A2H1VNW8 A0A2H1VP06 B0WB17 A0A2H1VPY0 A0A194Q004 A0A1Q3FPC2 A0A336LY62 A0A2H1VP00 A0A336LZS5 A0A2A4JKI4 A0A067QS64 A0A0N8P0I5 B3MI17 A0A1Q3FPF7 A0A1L8DE98 A0A1W4UP57 A0A2J7Q377 A0A2A4JKL3 Q17LW3 A0A1J1IVE3 A0A2J7Q364 A0A2M4CLX1 A0A0K8VIK3 A0A182LMR0 Q5BIE4 A0A0K8UFD6 A0A182Q5D9 A0A1S4GZN6 Q7JRA7 A0A182XNS8 A0A182U715 A0A2M4AF33 A0A0J9RHI1 B4NSL6 A0A034V4D0 A0A182VKS3 A0A182FN64 A0A194RFB6 W8AL50 W8AX47 A0A182Y0D9 W5JTB8 A0A182GLJ7 A0A2A4JRS5 A0A2M4AFD6 B4P485 A0A182I7D3 A0A194RFZ7 B3NKB8 A0A182K1W8 A0A0R1DS37 A0A0Q5VM65 B4HP21 A0A194PYB2 A0A182RI61 A0A212FKV3 A0A182PDV6 A0A0A1XJ44 A0A182MZ46 A0A0A1XHS7 A0A182W6C0 Q292H7 A0A2M3ZEY8 A0A2M4BLU2 T1PF36 A0A3B0JJR4 A0A1A9VA36 B4GDI7 A0A034VUA9 A0A1A9XAR9 A0A0R3NQT8 A0A1B0BN67 B4P486 A0A2A4JIL8 A0A2M4BMD9 A0A0L7R1A4 A0A151XIZ3 B3NKB7 A0A1A9WYG2 A0A2M4BMA3 A0A0A1XMC1 A0A182JDX2 A0A212FBF2 A0A0J9RG84
Pubmed
EMBL
BABH01032707
BABH01032708
BABH01032709
BABH01032710
BABH01032711
KZ150053
+ More
PZC74367.1 NWSH01000795 PCG74168.1 ODYU01005697 SOQ46811.1 ODYU01000213 SOQ34607.1 KQ460470 KPJ14575.1 KQ459585 KPI98329.1 RSAL01000225 RVE43977.1 PZC74368.1 PZC74369.1 KPI98328.1 KPJ14574.1 ODYU01013024 SOQ59639.1 ODYU01003582 SOQ42526.1 NWSH01001263 PCG71941.1 ODYU01003583 SOQ42529.1 SOQ42527.1 DS231876 EDS41975.1 ODYU01003747 SOQ42903.1 KPI98324.1 GFDL01005671 JAV29374.1 UFQS01000292 UFQT01000292 SSX02540.1 SSX22914.1 SOQ42528.1 UFQS01000360 UFQT01000360 SSX03128.1 SSX23494.1 PCG71943.1 KK853091 KDR11545.1 CH902619 KPU77220.1 EDV38027.1 GFDL01005653 JAV29392.1 GFDF01009316 JAV04768.1 NEVH01019069 PNF23027.1 PCG71942.1 CH477210 EAT47708.1 CVRI01000061 CRL04181.1 PNF23028.1 GGFL01002003 MBW66181.1 GDHF01029444 GDHF01016282 GDHF01013939 JAI22870.1 JAI36032.1 JAI38375.1 AE013599 BT021280 AAF57633.1 AAX33428.1 GDHF01027028 JAI25286.1 AXCN02000794 AAAB01008984 BT001852 AAF57634.1 AAN71613.1 GGFK01005927 MBW39248.1 CM002911 KMY94944.1 CH982331 EDX15594.1 KMY94943.1 GAKP01020827 GAKP01020826 JAC38126.1 KQ460211 KPJ16523.1 GAMC01017148 GAMC01017145 JAB89410.1 GAMC01017147 GAMC01017146 JAB89408.1 ADMH02000203 ETN67381.1 JXUM01072235 JXUM01072236 KQ562709 KXJ75293.1 NWSH01000802 PCG74112.1 GGFK01006136 MBW39457.1 CM000158 EDW91571.1 APCN01003380 KPJ16522.1 CH954179 EDV55140.2 KRJ99964.1 KQS62108.1 CH480816 EDW48523.1 KPI98326.1 AGBW02007960 OWR54367.1 GBXI01003739 JAD10553.1 GBXI01003780 JAD10512.1 CM000071 EAL24885.3 GGFM01006318 MBW27069.1 GGFJ01004823 MBW53964.1 KA647304 AFP61933.1 OUUW01000001 SPP73679.1 CH479181 EDW31644.1 GAKP01013839 JAC45113.1 KRT01647.1 JXJN01017213 EDW91572.1 PCG71945.1 GGFJ01004982 MBW54123.1 KQ414668 KOC64622.1 KQ982080 KYQ60190.1 EDV55139.1 GGFJ01004983 MBW54124.1 GBXI01004017 GBXI01002141 JAD10275.1 JAD12151.1 AGBW02009333 OWR51071.1 KMY94942.1
PZC74367.1 NWSH01000795 PCG74168.1 ODYU01005697 SOQ46811.1 ODYU01000213 SOQ34607.1 KQ460470 KPJ14575.1 KQ459585 KPI98329.1 RSAL01000225 RVE43977.1 PZC74368.1 PZC74369.1 KPI98328.1 KPJ14574.1 ODYU01013024 SOQ59639.1 ODYU01003582 SOQ42526.1 NWSH01001263 PCG71941.1 ODYU01003583 SOQ42529.1 SOQ42527.1 DS231876 EDS41975.1 ODYU01003747 SOQ42903.1 KPI98324.1 GFDL01005671 JAV29374.1 UFQS01000292 UFQT01000292 SSX02540.1 SSX22914.1 SOQ42528.1 UFQS01000360 UFQT01000360 SSX03128.1 SSX23494.1 PCG71943.1 KK853091 KDR11545.1 CH902619 KPU77220.1 EDV38027.1 GFDL01005653 JAV29392.1 GFDF01009316 JAV04768.1 NEVH01019069 PNF23027.1 PCG71942.1 CH477210 EAT47708.1 CVRI01000061 CRL04181.1 PNF23028.1 GGFL01002003 MBW66181.1 GDHF01029444 GDHF01016282 GDHF01013939 JAI22870.1 JAI36032.1 JAI38375.1 AE013599 BT021280 AAF57633.1 AAX33428.1 GDHF01027028 JAI25286.1 AXCN02000794 AAAB01008984 BT001852 AAF57634.1 AAN71613.1 GGFK01005927 MBW39248.1 CM002911 KMY94944.1 CH982331 EDX15594.1 KMY94943.1 GAKP01020827 GAKP01020826 JAC38126.1 KQ460211 KPJ16523.1 GAMC01017148 GAMC01017145 JAB89410.1 GAMC01017147 GAMC01017146 JAB89408.1 ADMH02000203 ETN67381.1 JXUM01072235 JXUM01072236 KQ562709 KXJ75293.1 NWSH01000802 PCG74112.1 GGFK01006136 MBW39457.1 CM000158 EDW91571.1 APCN01003380 KPJ16522.1 CH954179 EDV55140.2 KRJ99964.1 KQS62108.1 CH480816 EDW48523.1 KPI98326.1 AGBW02007960 OWR54367.1 GBXI01003739 JAD10553.1 GBXI01003780 JAD10512.1 CM000071 EAL24885.3 GGFM01006318 MBW27069.1 GGFJ01004823 MBW53964.1 KA647304 AFP61933.1 OUUW01000001 SPP73679.1 CH479181 EDW31644.1 GAKP01013839 JAC45113.1 KRT01647.1 JXJN01017213 EDW91572.1 PCG71945.1 GGFJ01004982 MBW54123.1 KQ414668 KOC64622.1 KQ982080 KYQ60190.1 EDV55139.1 GGFJ01004983 MBW54124.1 GBXI01004017 GBXI01002141 JAD10275.1 JAD12151.1 AGBW02009333 OWR51071.1 KMY94942.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000283053
UP000002320
+ More
UP000027135 UP000007801 UP000192221 UP000235965 UP000008820 UP000183832 UP000075882 UP000000803 UP000075886 UP000076407 UP000075902 UP000000304 UP000075903 UP000069272 UP000076408 UP000000673 UP000069940 UP000249989 UP000002282 UP000075840 UP000008711 UP000075881 UP000001292 UP000075900 UP000007151 UP000075885 UP000075884 UP000075920 UP000001819 UP000095301 UP000268350 UP000078200 UP000008744 UP000092443 UP000092460 UP000053825 UP000075809 UP000091820 UP000075880
UP000027135 UP000007801 UP000192221 UP000235965 UP000008820 UP000183832 UP000075882 UP000000803 UP000075886 UP000076407 UP000075902 UP000000304 UP000075903 UP000069272 UP000076408 UP000000673 UP000069940 UP000249989 UP000002282 UP000075840 UP000008711 UP000075881 UP000001292 UP000075900 UP000007151 UP000075885 UP000075884 UP000075920 UP000001819 UP000095301 UP000268350 UP000078200 UP000008744 UP000092443 UP000092460 UP000053825 UP000075809 UP000091820 UP000075880
Pfam
PF07690 MFS_1
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9JXJ3
A0A2W1BM66
A0A2A4JR34
A0A2H1W147
A0A2H1V191
A0A0N0PCS9
+ More
A0A194Q007 A0A3S2LCW7 A0A2W1BHP9 A0A2W1BGX7 A0A194PZ61 A0A0N1IEP7 A0A2H1X2V7 A0A2H1VNZ8 A0A2A4JIZ6 A0A2H1VNW8 A0A2H1VP06 B0WB17 A0A2H1VPY0 A0A194Q004 A0A1Q3FPC2 A0A336LY62 A0A2H1VP00 A0A336LZS5 A0A2A4JKI4 A0A067QS64 A0A0N8P0I5 B3MI17 A0A1Q3FPF7 A0A1L8DE98 A0A1W4UP57 A0A2J7Q377 A0A2A4JKL3 Q17LW3 A0A1J1IVE3 A0A2J7Q364 A0A2M4CLX1 A0A0K8VIK3 A0A182LMR0 Q5BIE4 A0A0K8UFD6 A0A182Q5D9 A0A1S4GZN6 Q7JRA7 A0A182XNS8 A0A182U715 A0A2M4AF33 A0A0J9RHI1 B4NSL6 A0A034V4D0 A0A182VKS3 A0A182FN64 A0A194RFB6 W8AL50 W8AX47 A0A182Y0D9 W5JTB8 A0A182GLJ7 A0A2A4JRS5 A0A2M4AFD6 B4P485 A0A182I7D3 A0A194RFZ7 B3NKB8 A0A182K1W8 A0A0R1DS37 A0A0Q5VM65 B4HP21 A0A194PYB2 A0A182RI61 A0A212FKV3 A0A182PDV6 A0A0A1XJ44 A0A182MZ46 A0A0A1XHS7 A0A182W6C0 Q292H7 A0A2M3ZEY8 A0A2M4BLU2 T1PF36 A0A3B0JJR4 A0A1A9VA36 B4GDI7 A0A034VUA9 A0A1A9XAR9 A0A0R3NQT8 A0A1B0BN67 B4P486 A0A2A4JIL8 A0A2M4BMD9 A0A0L7R1A4 A0A151XIZ3 B3NKB7 A0A1A9WYG2 A0A2M4BMA3 A0A0A1XMC1 A0A182JDX2 A0A212FBF2 A0A0J9RG84
A0A194Q007 A0A3S2LCW7 A0A2W1BHP9 A0A2W1BGX7 A0A194PZ61 A0A0N1IEP7 A0A2H1X2V7 A0A2H1VNZ8 A0A2A4JIZ6 A0A2H1VNW8 A0A2H1VP06 B0WB17 A0A2H1VPY0 A0A194Q004 A0A1Q3FPC2 A0A336LY62 A0A2H1VP00 A0A336LZS5 A0A2A4JKI4 A0A067QS64 A0A0N8P0I5 B3MI17 A0A1Q3FPF7 A0A1L8DE98 A0A1W4UP57 A0A2J7Q377 A0A2A4JKL3 Q17LW3 A0A1J1IVE3 A0A2J7Q364 A0A2M4CLX1 A0A0K8VIK3 A0A182LMR0 Q5BIE4 A0A0K8UFD6 A0A182Q5D9 A0A1S4GZN6 Q7JRA7 A0A182XNS8 A0A182U715 A0A2M4AF33 A0A0J9RHI1 B4NSL6 A0A034V4D0 A0A182VKS3 A0A182FN64 A0A194RFB6 W8AL50 W8AX47 A0A182Y0D9 W5JTB8 A0A182GLJ7 A0A2A4JRS5 A0A2M4AFD6 B4P485 A0A182I7D3 A0A194RFZ7 B3NKB8 A0A182K1W8 A0A0R1DS37 A0A0Q5VM65 B4HP21 A0A194PYB2 A0A182RI61 A0A212FKV3 A0A182PDV6 A0A0A1XJ44 A0A182MZ46 A0A0A1XHS7 A0A182W6C0 Q292H7 A0A2M3ZEY8 A0A2M4BLU2 T1PF36 A0A3B0JJR4 A0A1A9VA36 B4GDI7 A0A034VUA9 A0A1A9XAR9 A0A0R3NQT8 A0A1B0BN67 B4P486 A0A2A4JIL8 A0A2M4BMD9 A0A0L7R1A4 A0A151XIZ3 B3NKB7 A0A1A9WYG2 A0A2M4BMA3 A0A0A1XMC1 A0A182JDX2 A0A212FBF2 A0A0J9RG84
PDB
6E9O
E-value=5.66673e-13,
Score=181
Ontologies
GO
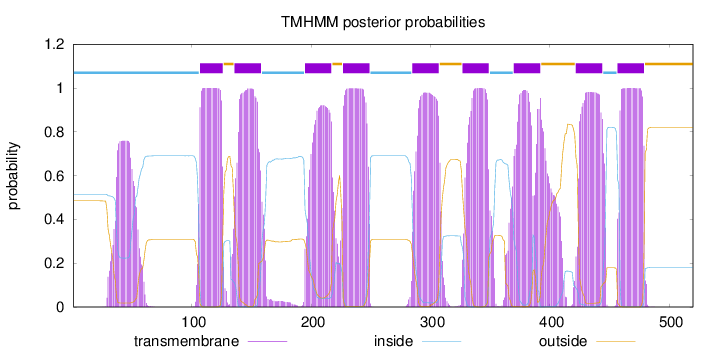
Topology
Length:
520
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
222.40624
Exp number, first 60 AAs:
15.40338
Total prob of N-in:
0.51498
POSSIBLE N-term signal
sequence
inside
1 - 106
TMhelix
107 - 126
outside
127 - 135
TMhelix
136 - 158
inside
159 - 194
TMhelix
195 - 217
outside
218 - 226
TMhelix
227 - 249
inside
250 - 284
TMhelix
285 - 307
outside
308 - 326
TMhelix
327 - 349
inside
350 - 369
TMhelix
370 - 392
outside
393 - 421
TMhelix
422 - 444
inside
445 - 456
TMhelix
457 - 479
outside
480 - 520
Population Genetic Test Statistics
Pi
17.334869
Theta
159.582288
Tajima's D
0.42474
CLR
0.476647
CSRT
0.493275336233188
Interpretation
Uncertain
