Gene
KWMTBOMO04236
Annotation
PREDICTED:_putative_inorganic_phosphate_cotransporter_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.932
Sequence
CDS
ATGGGAATGGCACATAGTTGCTTCTCCACTGTTTACGAGACCTTGATAGAACGATGGCTACCACCGAACGAAAGGAATACTTACAGTCATTTTATTTATGGAGGTATACAATTAGGAATCATAATAGGGCTACCAATCTCTGGTTTACTGTCTCAAACACGTCTTGGCTGGGAATTTATTTACTACGCCTTGGCCATGGCGACTTTGTCTATGGCAGTAGTTATAGGGACACTGACTGCTAGCTCATTTGAAGGACATCAGGCTATGGGGGATGGTGAAAAAGAGTTTATAAGAGACGCTATGACTTTGTATAAAAAGAAAAAACTGATTAGACCTTTTAGATCTATATTGGAATGCAAAAAGTTCTGGGCTGTGACAGTAGCTCACTCAGCGAGCAATGCTTTATTTATTTTTTACTTGATCTTCGTTCCTGTCTTCTTGGAAACGCTAAATGTTTCTCTTAAAGATGCTACCTTTTACACTATGATGTCATTTCTGACAATGTGGTTGGTTTACTTAACGACTTCTCCTACAGTAGAGTGGGTGGCTGGCTCCGAAATCGTAAACTATATTATGAGTACTACATGTCTCAGGAAAATAATTAACACCTTGGGTGCCCTCGGTATCATAATAGGTTTATCGATTCTACCCAATTTAACGCAGGAACGGACTAACGTGGCAGTTATTGTATTAATAGCAATATTAGGATTGCTTGGCTTCCAATTAACCGGTTTTTTGAGTAATTATCAAGACATGACAGAAAACTACAGTGGGACCCTCATAATGCTAAGCAGCACTATCAGCGGCTTCGTAGGAGCTGTCATCCCATTCCTATTCAGCTTTTTCTTGTATACAGAGCCGATCCATCTCTCTCAATGGAAAGCGGCGTTCTATATATTGGCAGCGTATTATATGGGATGCAATGCTGTTTATGTTTTATTCGCGAGCAATAAGCGACAACCGTGGGACGGTAACTCTGGCAGGAAAACGGGACATTACAACCACAGCTACAGCTATGACTCCAATCACATTCAACTCGAGGAGTTTTCACACTCAAAACTTATACAAGAGGAAAACGGTGCAATGATTTAA
Protein
MGMAHSCFSTVYETLIERWLPPNERNTYSHFIYGGIQLGIIIGLPISGLLSQTRLGWEFIYYALAMATLSMAVVIGTLTASSFEGHQAMGDGEKEFIRDAMTLYKKKKLIRPFRSILECKKFWAVTVAHSASNALFIFYLIFVPVFLETLNVSLKDATFYTMMSFLTMWLVYLTTSPTVEWVAGSEIVNYIMSTTCLRKIINTLGALGIIIGLSILPNLTQERTNVAVIVLIAILGLLGFQLTGFLSNYQDMTENYSGTLIMLSSTISGFVGAVIPFLFSFFLYTEPIHLSQWKAAFYILAAYYMGCNAVYVLFASNKRQPWDGNSGRKTGHYNHSYSYDSNHIQLEEFSHSKLIQEENGAMI
Summary
Uniprot
A0A2A4JQT1
A0A2W1BNC1
A0A2H1WPS7
A0A212F7R9
A0A0N0PD34
A0A194Q4J5
+ More
A0A0N1PJ27 A0A2W1BL17 A0A194PYB6 A0A2H1WFA7 A0A2A4JRY5 A0A3S2LT92 A0A212F7S2 A0A1Y1MXE7 A0A1Y1MXD1 E2BGX2 N6UML2 A0A194QFW7 A0A3L8E1X7 A0A026X3H4 U4TW53 A0A232EHZ9 K7IRU9 D6WYK6 A0A2R7W2R0 K7IRT8 A0A1B6E523 A0A0T6BG30 A0A2H1X2V7 A0A0T6AUS9 J9JXY7 X1WJ27 A0A154PQW6 A0A2H8TWW5 A0A2H8TIN5 A0A2A4JKL3 A0A1L8DE72 A0A1B6F5L1 D6WG71 A0A2S2PYP0 A0A195E864 A0A1L8DEJ4 E2AE42 A0A1Y1MVG1 A0A1Y1MRV4 F4WEN8 Q17LW3 A0A2H1VP06 A0A2H1VPY0 A0A2H1VNW8 A0A0N0U5C3 A0A0N0PCS9 A0A158NF06 A0A195AVS4 A0A2W1BHP9 A0A1B6GKY5 A0A1B6EWM7 A0A2S2PP82 A0A194Q007 A0A2A4JIZ6 A0A2H8TKU0 A0A195FLP4 A0A0L0CNZ5 A0A1B6M806 F4WEN7 A0A195E907 E1ZVK5 A0A1B6KZQ5 F4W6I0 A0A195C4Y5 A0A195C484 A0A0L7R1A4 A0A195FIV2 A0A0A1XMC1 A0A1B6F2P7 A0A0K8U2N6 A0A1B6KJM2 B3MJ87 A0A182JDX2 A0A2A4JKI4 A0A195D309 A0A158NF07 A0A2H1VP00 A0A151X227 A0A195AVB3 A0A1B6HF16 A0A151XIN9 A0A0A1XLG1 W8CD38 A0A195E4U7 A0A158P287 A0A151XIZ3 A0A1J1IVE3 A0A1W4VBM2 A0A0M5IXG7 A0A2T7P123
A0A0N1PJ27 A0A2W1BL17 A0A194PYB6 A0A2H1WFA7 A0A2A4JRY5 A0A3S2LT92 A0A212F7S2 A0A1Y1MXE7 A0A1Y1MXD1 E2BGX2 N6UML2 A0A194QFW7 A0A3L8E1X7 A0A026X3H4 U4TW53 A0A232EHZ9 K7IRU9 D6WYK6 A0A2R7W2R0 K7IRT8 A0A1B6E523 A0A0T6BG30 A0A2H1X2V7 A0A0T6AUS9 J9JXY7 X1WJ27 A0A154PQW6 A0A2H8TWW5 A0A2H8TIN5 A0A2A4JKL3 A0A1L8DE72 A0A1B6F5L1 D6WG71 A0A2S2PYP0 A0A195E864 A0A1L8DEJ4 E2AE42 A0A1Y1MVG1 A0A1Y1MRV4 F4WEN8 Q17LW3 A0A2H1VP06 A0A2H1VPY0 A0A2H1VNW8 A0A0N0U5C3 A0A0N0PCS9 A0A158NF06 A0A195AVS4 A0A2W1BHP9 A0A1B6GKY5 A0A1B6EWM7 A0A2S2PP82 A0A194Q007 A0A2A4JIZ6 A0A2H8TKU0 A0A195FLP4 A0A0L0CNZ5 A0A1B6M806 F4WEN7 A0A195E907 E1ZVK5 A0A1B6KZQ5 F4W6I0 A0A195C4Y5 A0A195C484 A0A0L7R1A4 A0A195FIV2 A0A0A1XMC1 A0A1B6F2P7 A0A0K8U2N6 A0A1B6KJM2 B3MJ87 A0A182JDX2 A0A2A4JKI4 A0A195D309 A0A158NF07 A0A2H1VP00 A0A151X227 A0A195AVB3 A0A1B6HF16 A0A151XIN9 A0A0A1XLG1 W8CD38 A0A195E4U7 A0A158P287 A0A151XIZ3 A0A1J1IVE3 A0A1W4VBM2 A0A0M5IXG7 A0A2T7P123
Pubmed
EMBL
NWSH01000795
PCG74169.1
KZ150053
PZC74366.1
ODYU01010152
SOQ55069.1
+ More
AGBW02009834 OWR49786.1 KQ460470 KPJ14576.1 KQ459585 KPI98330.1 KPJ14577.1 PZC74365.1 KPI98331.1 ODYU01005491 ODYU01008171 SOQ46394.1 SOQ51552.1 PCG74170.1 RSAL01000225 RVE43975.1 OWR49785.1 GEZM01018330 JAV90323.1 GEZM01018331 GEZM01018329 JAV90322.1 GL448228 EFN85104.1 APGK01017174 APGK01017175 APGK01017176 APGK01017177 KB739998 ENN81941.1 KQ459249 KPJ02341.1 QOIP01000002 RLU26028.1 KK107017 EZA62822.1 KB631278 ERL84203.1 NNAY01004388 OXU17964.1 KQ971362 EFA07863.1 KK854272 PTY14044.1 GEDC01004266 JAS33032.1 LJIG01000640 KRT86300.1 ODYU01013024 SOQ59639.1 LJIG01022770 KRT78806.1 ABLF02038141 ABLF02038142 ABLF02038149 ABLF02038153 ABLF02016022 ABLF02016025 ABLF02016026 KQ435056 KZC14291.1 GFXV01006605 MBW18410.1 GFXV01002171 MBW13976.1 NWSH01001263 PCG71942.1 GFDF01009417 JAV04667.1 GECZ01024227 JAS45542.1 KQ971320 EFA00540.2 GGMS01001435 MBY70638.1 KQ979479 KYN21311.1 GFDF01009314 JAV04770.1 GL438827 EFN68275.1 GEZM01023359 JAV88435.1 GEZM01023360 JAV88434.1 GL888103 EGI67401.1 CH477210 EAT47708.1 ODYU01003582 SOQ42527.1 ODYU01003747 SOQ42903.1 ODYU01003583 SOQ42529.1 KQ435783 KOX74621.1 KPJ14575.1 ADTU01013693 ADTU01013694 KQ976736 KYM76069.1 PZC74368.1 GECZ01006691 JAS63078.1 GECZ01027385 JAS42384.1 GGMR01018662 MBY31281.1 KPI98329.1 PCG71941.1 GFXV01002950 MBW14755.1 KQ981490 KYN41338.1 JRES01000105 KNC34073.1 GEBQ01007918 JAT32059.1 EGI67400.1 KYN21312.1 GL434548 EFN74774.1 GEBQ01023034 JAT16943.1 GL887707 EGI70315.1 KQ978287 KYM95665.1 KYM95664.1 KQ414668 KOC64622.1 KQ981523 KYN40328.1 GBXI01004017 GBXI01002141 JAD10275.1 JAD12151.1 GECZ01025331 JAS44438.1 GDHF01031536 JAI20778.1 GEBQ01028324 JAT11653.1 CH902619 EDV38181.2 PCG71943.1 KQ976885 KYN07282.1 ADTU01013695 SOQ42528.1 KQ982580 KYQ54483.1 KQ976731 KYM76178.1 GECU01034517 GECU01027976 JAS73189.1 JAS79730.1 KQ982080 KYQ60191.1 GBXI01002088 JAD12204.1 GAMC01002621 JAC03935.1 KQ979657 KYN19927.1 ADTU01007024 ADTU01007025 ADTU01007026 ADTU01007027 ADTU01007028 KYQ60190.1 CVRI01000061 CRL04181.1 CP012524 ALC41268.1 PZQS01000007 PVD27121.1
AGBW02009834 OWR49786.1 KQ460470 KPJ14576.1 KQ459585 KPI98330.1 KPJ14577.1 PZC74365.1 KPI98331.1 ODYU01005491 ODYU01008171 SOQ46394.1 SOQ51552.1 PCG74170.1 RSAL01000225 RVE43975.1 OWR49785.1 GEZM01018330 JAV90323.1 GEZM01018331 GEZM01018329 JAV90322.1 GL448228 EFN85104.1 APGK01017174 APGK01017175 APGK01017176 APGK01017177 KB739998 ENN81941.1 KQ459249 KPJ02341.1 QOIP01000002 RLU26028.1 KK107017 EZA62822.1 KB631278 ERL84203.1 NNAY01004388 OXU17964.1 KQ971362 EFA07863.1 KK854272 PTY14044.1 GEDC01004266 JAS33032.1 LJIG01000640 KRT86300.1 ODYU01013024 SOQ59639.1 LJIG01022770 KRT78806.1 ABLF02038141 ABLF02038142 ABLF02038149 ABLF02038153 ABLF02016022 ABLF02016025 ABLF02016026 KQ435056 KZC14291.1 GFXV01006605 MBW18410.1 GFXV01002171 MBW13976.1 NWSH01001263 PCG71942.1 GFDF01009417 JAV04667.1 GECZ01024227 JAS45542.1 KQ971320 EFA00540.2 GGMS01001435 MBY70638.1 KQ979479 KYN21311.1 GFDF01009314 JAV04770.1 GL438827 EFN68275.1 GEZM01023359 JAV88435.1 GEZM01023360 JAV88434.1 GL888103 EGI67401.1 CH477210 EAT47708.1 ODYU01003582 SOQ42527.1 ODYU01003747 SOQ42903.1 ODYU01003583 SOQ42529.1 KQ435783 KOX74621.1 KPJ14575.1 ADTU01013693 ADTU01013694 KQ976736 KYM76069.1 PZC74368.1 GECZ01006691 JAS63078.1 GECZ01027385 JAS42384.1 GGMR01018662 MBY31281.1 KPI98329.1 PCG71941.1 GFXV01002950 MBW14755.1 KQ981490 KYN41338.1 JRES01000105 KNC34073.1 GEBQ01007918 JAT32059.1 EGI67400.1 KYN21312.1 GL434548 EFN74774.1 GEBQ01023034 JAT16943.1 GL887707 EGI70315.1 KQ978287 KYM95665.1 KYM95664.1 KQ414668 KOC64622.1 KQ981523 KYN40328.1 GBXI01004017 GBXI01002141 JAD10275.1 JAD12151.1 GECZ01025331 JAS44438.1 GDHF01031536 JAI20778.1 GEBQ01028324 JAT11653.1 CH902619 EDV38181.2 PCG71943.1 KQ976885 KYN07282.1 ADTU01013695 SOQ42528.1 KQ982580 KYQ54483.1 KQ976731 KYM76178.1 GECU01034517 GECU01027976 JAS73189.1 JAS79730.1 KQ982080 KYQ60191.1 GBXI01002088 JAD12204.1 GAMC01002621 JAC03935.1 KQ979657 KYN19927.1 ADTU01007024 ADTU01007025 ADTU01007026 ADTU01007027 ADTU01007028 KYQ60190.1 CVRI01000061 CRL04181.1 CP012524 ALC41268.1 PZQS01000007 PVD27121.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000283053
UP000008237
+ More
UP000019118 UP000279307 UP000053097 UP000030742 UP000215335 UP000002358 UP000007266 UP000007819 UP000076502 UP000078492 UP000000311 UP000007755 UP000008820 UP000053105 UP000005205 UP000078540 UP000078541 UP000037069 UP000078542 UP000053825 UP000007801 UP000075880 UP000075809 UP000183832 UP000192221 UP000092553 UP000245119
UP000019118 UP000279307 UP000053097 UP000030742 UP000215335 UP000002358 UP000007266 UP000007819 UP000076502 UP000078492 UP000000311 UP000007755 UP000008820 UP000053105 UP000005205 UP000078540 UP000078541 UP000037069 UP000078542 UP000053825 UP000007801 UP000075880 UP000075809 UP000183832 UP000192221 UP000092553 UP000245119
Pfam
PF07690 MFS_1
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A2A4JQT1
A0A2W1BNC1
A0A2H1WPS7
A0A212F7R9
A0A0N0PD34
A0A194Q4J5
+ More
A0A0N1PJ27 A0A2W1BL17 A0A194PYB6 A0A2H1WFA7 A0A2A4JRY5 A0A3S2LT92 A0A212F7S2 A0A1Y1MXE7 A0A1Y1MXD1 E2BGX2 N6UML2 A0A194QFW7 A0A3L8E1X7 A0A026X3H4 U4TW53 A0A232EHZ9 K7IRU9 D6WYK6 A0A2R7W2R0 K7IRT8 A0A1B6E523 A0A0T6BG30 A0A2H1X2V7 A0A0T6AUS9 J9JXY7 X1WJ27 A0A154PQW6 A0A2H8TWW5 A0A2H8TIN5 A0A2A4JKL3 A0A1L8DE72 A0A1B6F5L1 D6WG71 A0A2S2PYP0 A0A195E864 A0A1L8DEJ4 E2AE42 A0A1Y1MVG1 A0A1Y1MRV4 F4WEN8 Q17LW3 A0A2H1VP06 A0A2H1VPY0 A0A2H1VNW8 A0A0N0U5C3 A0A0N0PCS9 A0A158NF06 A0A195AVS4 A0A2W1BHP9 A0A1B6GKY5 A0A1B6EWM7 A0A2S2PP82 A0A194Q007 A0A2A4JIZ6 A0A2H8TKU0 A0A195FLP4 A0A0L0CNZ5 A0A1B6M806 F4WEN7 A0A195E907 E1ZVK5 A0A1B6KZQ5 F4W6I0 A0A195C4Y5 A0A195C484 A0A0L7R1A4 A0A195FIV2 A0A0A1XMC1 A0A1B6F2P7 A0A0K8U2N6 A0A1B6KJM2 B3MJ87 A0A182JDX2 A0A2A4JKI4 A0A195D309 A0A158NF07 A0A2H1VP00 A0A151X227 A0A195AVB3 A0A1B6HF16 A0A151XIN9 A0A0A1XLG1 W8CD38 A0A195E4U7 A0A158P287 A0A151XIZ3 A0A1J1IVE3 A0A1W4VBM2 A0A0M5IXG7 A0A2T7P123
A0A0N1PJ27 A0A2W1BL17 A0A194PYB6 A0A2H1WFA7 A0A2A4JRY5 A0A3S2LT92 A0A212F7S2 A0A1Y1MXE7 A0A1Y1MXD1 E2BGX2 N6UML2 A0A194QFW7 A0A3L8E1X7 A0A026X3H4 U4TW53 A0A232EHZ9 K7IRU9 D6WYK6 A0A2R7W2R0 K7IRT8 A0A1B6E523 A0A0T6BG30 A0A2H1X2V7 A0A0T6AUS9 J9JXY7 X1WJ27 A0A154PQW6 A0A2H8TWW5 A0A2H8TIN5 A0A2A4JKL3 A0A1L8DE72 A0A1B6F5L1 D6WG71 A0A2S2PYP0 A0A195E864 A0A1L8DEJ4 E2AE42 A0A1Y1MVG1 A0A1Y1MRV4 F4WEN8 Q17LW3 A0A2H1VP06 A0A2H1VPY0 A0A2H1VNW8 A0A0N0U5C3 A0A0N0PCS9 A0A158NF06 A0A195AVS4 A0A2W1BHP9 A0A1B6GKY5 A0A1B6EWM7 A0A2S2PP82 A0A194Q007 A0A2A4JIZ6 A0A2H8TKU0 A0A195FLP4 A0A0L0CNZ5 A0A1B6M806 F4WEN7 A0A195E907 E1ZVK5 A0A1B6KZQ5 F4W6I0 A0A195C4Y5 A0A195C484 A0A0L7R1A4 A0A195FIV2 A0A0A1XMC1 A0A1B6F2P7 A0A0K8U2N6 A0A1B6KJM2 B3MJ87 A0A182JDX2 A0A2A4JKI4 A0A195D309 A0A158NF07 A0A2H1VP00 A0A151X227 A0A195AVB3 A0A1B6HF16 A0A151XIN9 A0A0A1XLG1 W8CD38 A0A195E4U7 A0A158P287 A0A151XIZ3 A0A1J1IVE3 A0A1W4VBM2 A0A0M5IXG7 A0A2T7P123
Ontologies
GO
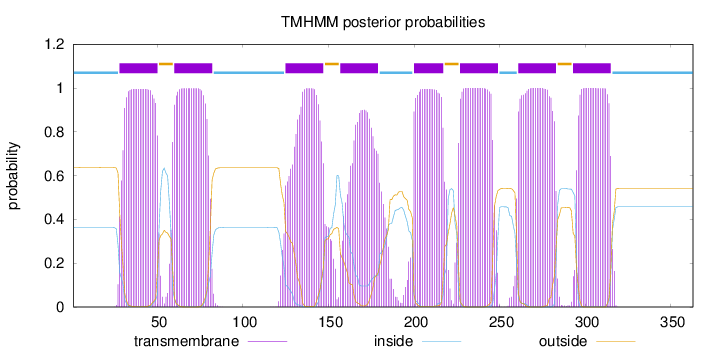
Topology
Length:
363
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
171.28054
Exp number, first 60 AAs:
24.10479
Total prob of N-in:
0.36253
POSSIBLE N-term signal
sequence
inside
1 - 27
TMhelix
28 - 50
outside
51 - 59
TMhelix
60 - 82
inside
83 - 124
TMhelix
125 - 147
outside
148 - 156
TMhelix
157 - 179
inside
180 - 199
TMhelix
200 - 217
outside
218 - 226
TMhelix
227 - 249
inside
250 - 260
TMhelix
261 - 283
outside
284 - 292
TMhelix
293 - 315
inside
316 - 363
Population Genetic Test Statistics
Pi
215.157857
Theta
24.194463
Tajima's D
0.549882
CLR
0.622978
CSRT
0.534873256337183
Interpretation
Uncertain
