Gene
KWMTBOMO04235
Pre Gene Modal
BGIBMGA014257
Annotation
PREDICTED:_putative_inorganic_phosphate_cotransporter_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.927
Sequence
CDS
ATGCAGATACCAGCTGGACTCCTCGCAAAGAAGTATGGGGGAAAACCAATACTGCTGGTCGCATTGTTAGCAAATGGGTCACTTTGCGTGCTGTTGCCAACGGTCGCTAGTTTGGGAGGTTGGCCGATGATATGCCTCTGTCGAGTACTCATGGGCTTAACGCAAGCTTGCCTCTTTCCAGCCAGTCACACGCTCCTGGGAAGATGGTTACCAGCAAACGAACGTACTACCCTGGCTGGCGTTGTATACGGAGGAGCACAAATGGGTACAATCATAGCCATGCCGGTATCAGGAGTTTTGGCAGAAACTGCAATGGGCTGGAAGTTAATCTTTTATGCTACGGGAGGATCTATGTTTGTCACTGCTGGACTCTGGTTCTGGTTTTCGGCCAGCTCTCCCGAAGAACATCCAATGATGTCTGAAGAAGAGAAATATTATATTGAAATGGGGTTGAATATGAGTACGGCCTTTCACATGGGCAACCGCCCGACACCATGGAAAGAGATGCTTAAGACAGGGCCACTGTGGGCCATCACGGTCTCGCATATCGCCAACACCGTCGCTTTCGTGCTATTTTTCGTCGACTTGCCAACGTACTTGGAAATGGCCATGCAAATATCTCTGAAGAATAGTGCTCTGCTATCAGCGCTACCTTATTTCGGTATGTGGGTTGGCAGTATTGCCTCTTCAATTATTTGTGAGAAGATATACAACAGAGGTTTACTTTCCATCGGCGCCTGTAGGAAGATATTCAATTCTTTAGGATTATTCGGGACGGGTGCGGGATTAATTGTCTTAGCTTTTATGGGGCCAGAACACAAGAATATAGCTATCGCCACTGTAGTTGCATTACTTACTATGTGTGGTTGCACTTCTGCCGGCTTCATGGTAAATCACTTGGACCTGTCTCCACACTTTGCCGGCGTGATGCTCTCTATAACCAACTTCGCCGGTAGTATCGGCAGTATACTAACGCCGGTTGGCACTAGTCTTATATTACGTAATGATTCGTCGGACATCTCAAGTTGGCGCATCGTATTTCTAGCAACCTCGGCCATTTGCGTCACTGCCAACATCATTTACCTGATATTCGGCAGTGGCGAGAGGCAGCCCTGGGATCGTCCTGATTGGGGAAACAGAAATAAATCAGATGCAGAAGAAATACGTCCGGTGTTGACTATGAAACATTTAAATGTTGAAGAACAGGAGAAGACTGCAGAACCATAG
Protein
MQIPAGLLAKKYGGKPILLVALLANGSLCVLLPTVASLGGWPMICLCRVLMGLTQACLFPASHTLLGRWLPANERTTLAGVVYGGAQMGTIIAMPVSGVLAETAMGWKLIFYATGGSMFVTAGLWFWFSASSPEEHPMMSEEEKYYIEMGLNMSTAFHMGNRPTPWKEMLKTGPLWAITVSHIANTVAFVLFFVDLPTYLEMAMQISLKNSALLSALPYFGMWVGSIASSIICEKIYNRGLLSIGACRKIFNSLGLFGTGAGLIVLAFMGPEHKNIAIATVVALLTMCGCTSAGFMVNHLDLSPHFAGVMLSITNFAGSIGSILTPVGTSLILRNDSSDISSWRIVFLATSAICVTANIIYLIFGSGERQPWDRPDWGNRNKSDAEEIRPVLTMKHLNVEEQEKTAEP
Summary
Uniprot
A0A2W1BL17
A0A2A4JRY5
A0A2H1WFA7
A0A194PYB6
A0A212F7S2
A0A0L7LQA2
+ More
A0A3S2LT92 A0A0N1PJ27 A0A2H1W184 A0A194Q004 A0A194RFB6 U5EXD9 A0A1J1IVE3 A0A0L7R1A3 A0A336LY62 A0A336LZS5 A0A212FBF2 A0A1Q3FPF7 A0A1Q3FPC2 A0A1L8DEB4 B0WB17 A0A088AJE0 A0A2J7Q364 A0A2J7Q377 A0A0Q5VM65 A0A0R1DS37 B4P485 A0A0K8U2N6 B3NKB8 A0A2A4JRS5 A0A2W1BRH8 U4TW53 A0A2A3E4U8 A0A0N8P0I5 N6UML2 A0A1L8DEJ4 A0A0A1XMC1 A0A1Q3FPK7 A0A1Q3FPK6 B3MI17 A0A2H1V268 A0A0B4K6S9 A0A2A4JQ34 A0A2W1BJ80 A0A0Q5WNR9 B4LJ26 Q8MS66 A0A1W4W9U6 A0A0J9R8U8 A0A0J9TVR6 B3N9C7 A0A1W4VYZ2 A0A0Q9W4Y3 X1WJ27 A0A182GLJ7 Q17LW3 B4HR96 A0A0Q5WBY5 B4NSL6 A0A2M4CLX1 A0A182FN64 A0A0Q9X7W0 Q7JRA7 A0A0L0CKT9 Q5BIE4 A0A1W4WJ07 A0A2H1V245 A0A0J9RHI1 B4KPC7 A0A2H1VD15 A0A0A1WUR6 B4HP21 A0A1I8NMZ7 W5JTB8 A0A2M3ZEY8 A0A0R1DM54 A0A0A1X1J1 A0A0M3QVP0 B4QEU3 A0A1I8NN03 A0A2M4BMD9 B4P254 A0A1B6E0U8 Q7PWK4 A0A2A4JIL8 A0A0R1DTB4 A0A182VKS3 A0A2M4BLU2 A0A1I8QEX4 A0A182Y0D9 A0A1L8E9W4 B4J858 A0A182I7D3 A0A2M4AF33 A0A067QS64 A0A084VL23 A0A1I8N0V7 A0A2M4BMC5 A0A1B6E1K5
A0A3S2LT92 A0A0N1PJ27 A0A2H1W184 A0A194Q004 A0A194RFB6 U5EXD9 A0A1J1IVE3 A0A0L7R1A3 A0A336LY62 A0A336LZS5 A0A212FBF2 A0A1Q3FPF7 A0A1Q3FPC2 A0A1L8DEB4 B0WB17 A0A088AJE0 A0A2J7Q364 A0A2J7Q377 A0A0Q5VM65 A0A0R1DS37 B4P485 A0A0K8U2N6 B3NKB8 A0A2A4JRS5 A0A2W1BRH8 U4TW53 A0A2A3E4U8 A0A0N8P0I5 N6UML2 A0A1L8DEJ4 A0A0A1XMC1 A0A1Q3FPK7 A0A1Q3FPK6 B3MI17 A0A2H1V268 A0A0B4K6S9 A0A2A4JQ34 A0A2W1BJ80 A0A0Q5WNR9 B4LJ26 Q8MS66 A0A1W4W9U6 A0A0J9R8U8 A0A0J9TVR6 B3N9C7 A0A1W4VYZ2 A0A0Q9W4Y3 X1WJ27 A0A182GLJ7 Q17LW3 B4HR96 A0A0Q5WBY5 B4NSL6 A0A2M4CLX1 A0A182FN64 A0A0Q9X7W0 Q7JRA7 A0A0L0CKT9 Q5BIE4 A0A1W4WJ07 A0A2H1V245 A0A0J9RHI1 B4KPC7 A0A2H1VD15 A0A0A1WUR6 B4HP21 A0A1I8NMZ7 W5JTB8 A0A2M3ZEY8 A0A0R1DM54 A0A0A1X1J1 A0A0M3QVP0 B4QEU3 A0A1I8NN03 A0A2M4BMD9 B4P254 A0A1B6E0U8 Q7PWK4 A0A2A4JIL8 A0A0R1DTB4 A0A182VKS3 A0A2M4BLU2 A0A1I8QEX4 A0A182Y0D9 A0A1L8E9W4 B4J858 A0A182I7D3 A0A2M4AF33 A0A067QS64 A0A084VL23 A0A1I8N0V7 A0A2M4BMC5 A0A1B6E1K5
Pubmed
EMBL
KZ150053
PZC74365.1
NWSH01000795
PCG74170.1
ODYU01005491
ODYU01008171
+ More
SOQ46394.1 SOQ51552.1 KQ459585 KPI98331.1 AGBW02009834 OWR49785.1 JTDY01000396 KOB77376.1 RSAL01000225 RVE43975.1 KQ460470 KPJ14577.1 ODYU01005717 SOQ46839.1 KPI98324.1 KQ460211 KPJ16523.1 GANO01002583 JAB57288.1 CVRI01000061 CRL04181.1 KQ414668 KOC64623.1 UFQS01000292 UFQT01000292 SSX02540.1 SSX22914.1 UFQS01000360 UFQT01000360 SSX03128.1 SSX23494.1 AGBW02009333 OWR51071.1 GFDL01005653 JAV29392.1 GFDL01005671 JAV29374.1 GFDF01009281 JAV04803.1 DS231876 EDS41975.1 NEVH01019069 PNF23028.1 PNF23027.1 CH954179 KQS62108.1 CM000158 KRJ99964.1 EDW91571.1 GDHF01031536 JAI20778.1 EDV55140.2 NWSH01000802 PCG74112.1 PZC74363.1 KB631278 ERL84203.1 KZ288409 PBC26101.1 CH902619 KPU77220.1 APGK01017174 APGK01017175 APGK01017176 APGK01017177 KB739998 ENN81941.1 GFDF01009314 JAV04770.1 GBXI01004017 GBXI01002141 JAD10275.1 JAD12151.1 GFDL01005603 JAV29442.1 GFDL01005627 JAV29418.1 EDV38027.1 ODYU01000335 SOQ34909.1 AE013599 AFH07950.1 AFH07951.1 PCG74111.1 PZC74361.1 CH954177 KQS70911.1 CH940648 EDW61462.1 AY119060 AAF59171.3 AAM50920.1 CM002911 KMY92119.1 KMY92120.1 EDV59614.1 KRF79997.1 ABLF02016022 ABLF02016025 ABLF02016026 JXUM01072235 JXUM01072236 KQ562709 KXJ75293.1 CH477210 EAT47708.1 CH480816 EDW46839.1 KQS70912.1 CH982331 EDX15594.1 KMY94943.1 GGFL01002003 MBW66181.1 CH933808 KRG04447.1 BT001852 AAF57634.1 AAN71613.1 JRES01000264 KNC32870.1 BT021280 AAF57633.1 AAX33428.1 ODYU01000336 SOQ34910.1 KMY94944.1 EDW09103.1 ODYU01001711 SOQ38292.1 GBXI01012109 JAD02183.1 EDW48523.1 ADMH02000203 ETN67381.1 GGFM01006318 MBW27069.1 CM000157 KRJ98328.1 GBXI01009556 JAD04736.1 CP012524 ALC42739.1 CM000362 EDX06089.1 GGFJ01004982 MBW54123.1 EDW89255.1 GEDC01005745 JAS31553.1 AAAB01008984 EAA14814.5 NWSH01001263 PCG71945.1 KRJ98329.1 GGFJ01004823 MBW53964.1 GFDG01003272 JAV15527.1 CH916367 EDW01195.1 APCN01003380 GGFK01005927 MBW39248.1 KK853091 KDR11545.1 ATLV01014397 KE524970 KFB38667.1 GGFJ01005003 MBW54144.1 GEDC01005492 JAS31806.1
SOQ46394.1 SOQ51552.1 KQ459585 KPI98331.1 AGBW02009834 OWR49785.1 JTDY01000396 KOB77376.1 RSAL01000225 RVE43975.1 KQ460470 KPJ14577.1 ODYU01005717 SOQ46839.1 KPI98324.1 KQ460211 KPJ16523.1 GANO01002583 JAB57288.1 CVRI01000061 CRL04181.1 KQ414668 KOC64623.1 UFQS01000292 UFQT01000292 SSX02540.1 SSX22914.1 UFQS01000360 UFQT01000360 SSX03128.1 SSX23494.1 AGBW02009333 OWR51071.1 GFDL01005653 JAV29392.1 GFDL01005671 JAV29374.1 GFDF01009281 JAV04803.1 DS231876 EDS41975.1 NEVH01019069 PNF23028.1 PNF23027.1 CH954179 KQS62108.1 CM000158 KRJ99964.1 EDW91571.1 GDHF01031536 JAI20778.1 EDV55140.2 NWSH01000802 PCG74112.1 PZC74363.1 KB631278 ERL84203.1 KZ288409 PBC26101.1 CH902619 KPU77220.1 APGK01017174 APGK01017175 APGK01017176 APGK01017177 KB739998 ENN81941.1 GFDF01009314 JAV04770.1 GBXI01004017 GBXI01002141 JAD10275.1 JAD12151.1 GFDL01005603 JAV29442.1 GFDL01005627 JAV29418.1 EDV38027.1 ODYU01000335 SOQ34909.1 AE013599 AFH07950.1 AFH07951.1 PCG74111.1 PZC74361.1 CH954177 KQS70911.1 CH940648 EDW61462.1 AY119060 AAF59171.3 AAM50920.1 CM002911 KMY92119.1 KMY92120.1 EDV59614.1 KRF79997.1 ABLF02016022 ABLF02016025 ABLF02016026 JXUM01072235 JXUM01072236 KQ562709 KXJ75293.1 CH477210 EAT47708.1 CH480816 EDW46839.1 KQS70912.1 CH982331 EDX15594.1 KMY94943.1 GGFL01002003 MBW66181.1 CH933808 KRG04447.1 BT001852 AAF57634.1 AAN71613.1 JRES01000264 KNC32870.1 BT021280 AAF57633.1 AAX33428.1 ODYU01000336 SOQ34910.1 KMY94944.1 EDW09103.1 ODYU01001711 SOQ38292.1 GBXI01012109 JAD02183.1 EDW48523.1 ADMH02000203 ETN67381.1 GGFM01006318 MBW27069.1 CM000157 KRJ98328.1 GBXI01009556 JAD04736.1 CP012524 ALC42739.1 CM000362 EDX06089.1 GGFJ01004982 MBW54123.1 EDW89255.1 GEDC01005745 JAS31553.1 AAAB01008984 EAA14814.5 NWSH01001263 PCG71945.1 KRJ98329.1 GGFJ01004823 MBW53964.1 GFDG01003272 JAV15527.1 CH916367 EDW01195.1 APCN01003380 GGFK01005927 MBW39248.1 KK853091 KDR11545.1 ATLV01014397 KE524970 KFB38667.1 GGFJ01005003 MBW54144.1 GEDC01005492 JAS31806.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000037510
UP000283053
UP000053240
+ More
UP000183832 UP000053825 UP000002320 UP000005203 UP000235965 UP000008711 UP000002282 UP000030742 UP000242457 UP000007801 UP000019118 UP000000803 UP000008792 UP000192221 UP000007819 UP000069940 UP000249989 UP000008820 UP000001292 UP000000304 UP000069272 UP000009192 UP000037069 UP000192223 UP000095300 UP000000673 UP000092553 UP000007062 UP000075903 UP000076408 UP000001070 UP000075840 UP000027135 UP000030765 UP000095301
UP000183832 UP000053825 UP000002320 UP000005203 UP000235965 UP000008711 UP000002282 UP000030742 UP000242457 UP000007801 UP000019118 UP000000803 UP000008792 UP000192221 UP000007819 UP000069940 UP000249989 UP000008820 UP000001292 UP000000304 UP000069272 UP000009192 UP000037069 UP000192223 UP000095300 UP000000673 UP000092553 UP000007062 UP000075903 UP000076408 UP000001070 UP000075840 UP000027135 UP000030765 UP000095301
Interpro
ProteinModelPortal
A0A2W1BL17
A0A2A4JRY5
A0A2H1WFA7
A0A194PYB6
A0A212F7S2
A0A0L7LQA2
+ More
A0A3S2LT92 A0A0N1PJ27 A0A2H1W184 A0A194Q004 A0A194RFB6 U5EXD9 A0A1J1IVE3 A0A0L7R1A3 A0A336LY62 A0A336LZS5 A0A212FBF2 A0A1Q3FPF7 A0A1Q3FPC2 A0A1L8DEB4 B0WB17 A0A088AJE0 A0A2J7Q364 A0A2J7Q377 A0A0Q5VM65 A0A0R1DS37 B4P485 A0A0K8U2N6 B3NKB8 A0A2A4JRS5 A0A2W1BRH8 U4TW53 A0A2A3E4U8 A0A0N8P0I5 N6UML2 A0A1L8DEJ4 A0A0A1XMC1 A0A1Q3FPK7 A0A1Q3FPK6 B3MI17 A0A2H1V268 A0A0B4K6S9 A0A2A4JQ34 A0A2W1BJ80 A0A0Q5WNR9 B4LJ26 Q8MS66 A0A1W4W9U6 A0A0J9R8U8 A0A0J9TVR6 B3N9C7 A0A1W4VYZ2 A0A0Q9W4Y3 X1WJ27 A0A182GLJ7 Q17LW3 B4HR96 A0A0Q5WBY5 B4NSL6 A0A2M4CLX1 A0A182FN64 A0A0Q9X7W0 Q7JRA7 A0A0L0CKT9 Q5BIE4 A0A1W4WJ07 A0A2H1V245 A0A0J9RHI1 B4KPC7 A0A2H1VD15 A0A0A1WUR6 B4HP21 A0A1I8NMZ7 W5JTB8 A0A2M3ZEY8 A0A0R1DM54 A0A0A1X1J1 A0A0M3QVP0 B4QEU3 A0A1I8NN03 A0A2M4BMD9 B4P254 A0A1B6E0U8 Q7PWK4 A0A2A4JIL8 A0A0R1DTB4 A0A182VKS3 A0A2M4BLU2 A0A1I8QEX4 A0A182Y0D9 A0A1L8E9W4 B4J858 A0A182I7D3 A0A2M4AF33 A0A067QS64 A0A084VL23 A0A1I8N0V7 A0A2M4BMC5 A0A1B6E1K5
A0A3S2LT92 A0A0N1PJ27 A0A2H1W184 A0A194Q004 A0A194RFB6 U5EXD9 A0A1J1IVE3 A0A0L7R1A3 A0A336LY62 A0A336LZS5 A0A212FBF2 A0A1Q3FPF7 A0A1Q3FPC2 A0A1L8DEB4 B0WB17 A0A088AJE0 A0A2J7Q364 A0A2J7Q377 A0A0Q5VM65 A0A0R1DS37 B4P485 A0A0K8U2N6 B3NKB8 A0A2A4JRS5 A0A2W1BRH8 U4TW53 A0A2A3E4U8 A0A0N8P0I5 N6UML2 A0A1L8DEJ4 A0A0A1XMC1 A0A1Q3FPK7 A0A1Q3FPK6 B3MI17 A0A2H1V268 A0A0B4K6S9 A0A2A4JQ34 A0A2W1BJ80 A0A0Q5WNR9 B4LJ26 Q8MS66 A0A1W4W9U6 A0A0J9R8U8 A0A0J9TVR6 B3N9C7 A0A1W4VYZ2 A0A0Q9W4Y3 X1WJ27 A0A182GLJ7 Q17LW3 B4HR96 A0A0Q5WBY5 B4NSL6 A0A2M4CLX1 A0A182FN64 A0A0Q9X7W0 Q7JRA7 A0A0L0CKT9 Q5BIE4 A0A1W4WJ07 A0A2H1V245 A0A0J9RHI1 B4KPC7 A0A2H1VD15 A0A0A1WUR6 B4HP21 A0A1I8NMZ7 W5JTB8 A0A2M3ZEY8 A0A0R1DM54 A0A0A1X1J1 A0A0M3QVP0 B4QEU3 A0A1I8NN03 A0A2M4BMD9 B4P254 A0A1B6E0U8 Q7PWK4 A0A2A4JIL8 A0A0R1DTB4 A0A182VKS3 A0A2M4BLU2 A0A1I8QEX4 A0A182Y0D9 A0A1L8E9W4 B4J858 A0A182I7D3 A0A2M4AF33 A0A067QS64 A0A084VL23 A0A1I8N0V7 A0A2M4BMC5 A0A1B6E1K5
PDB
6E9O
E-value=2.48428e-07,
Score=131
Ontologies
GO
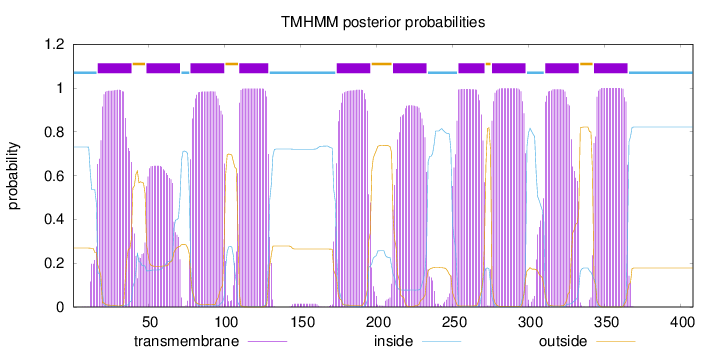
Topology
Length:
408
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
210.17675
Exp number, first 60 AAs:
31.40731
Total prob of N-in:
0.73078
POSSIBLE N-term signal
sequence
inside
1 - 16
TMhelix
17 - 39
outside
40 - 48
TMhelix
49 - 71
inside
72 - 77
TMhelix
78 - 100
outside
101 - 109
TMhelix
110 - 129
inside
130 - 173
TMhelix
174 - 196
outside
197 - 210
TMhelix
211 - 233
inside
234 - 253
TMhelix
254 - 271
outside
272 - 275
TMhelix
276 - 298
inside
299 - 310
TMhelix
311 - 333
outside
334 - 342
TMhelix
343 - 365
inside
366 - 408
Population Genetic Test Statistics
Pi
257.166392
Theta
171.040101
Tajima's D
0.754875
CLR
0.629943
CSRT
0.591220438978051
Interpretation
Uncertain
