Gene
KWMTBOMO04234
Pre Gene Modal
BGIBMGA014223
Annotation
Uncharacterized_protein_OBRU01_23542_[Operophtera_brumata]
Location in the cell
PlasmaMembrane Reliability : 4.855
Sequence
CDS
ATGGAATTGCAGGGAAAGTATTGCACGGTCCTCGTGCCCTGGGACCAGATGCCTACAAACCTACCAGAAACATTAGAAGATAACTGGTTACAGCTATTTCTGACCTATCAATTCCGGGGTATTTCTGGAATGCTGGAGCAAAATAAGGGATATGCTCATGAAGTGTCCAGTATAAGAGGACGCATTGACCCAAACGAGCATACAGTATTTGTGCTGAGTTCAAATGAAACTCTTCTAGGCCTTTGCATACCGAAAGTGTGTACGACATCGGAAGCGCTCAACCATGTTTTGAACTACATCAGCTTTGTCAATTTAACCTACAGGGATTATTACTGTCGGTTGCCGAATGACAAGCCTTTTAGACCCGCTGATTATGTATCTATGGCTATATTTTCAATTATAGCGATTTTAACAGTGGCAAGTTCTTCATATGAGATTTACAATGTCCACGTACTAAAAAAAGATTCATCACAGTTGCACCCTATTTATCGAAATTTCTCAGTCTTTCACAATACCCGCCGTTTCCTGACCTTCAACCGTAATTCTGGACAATTAGACTGTTTGGACGGTATAAGAGTGATGGCCATGTCATGGATTACTTTAGGCCACACTTTTTGCATGATCATCTTTGCTTTTACTCATAATAGTATATACAGAGTAATTAAGTTCAGGCAGCTAGAAACAAATGTTATCTTCGGTGCGATCCTGTCTGTTGATACATTCTTATCAATTAGCGGACTTCTACTTGTTTATACGGTTGTTGGGAAAATACCTAGAGATCGTTTTCTTAAAGGGATCCCATTGTTTTATTTATACCGATTATTAAGAATATGGCCGCTTCTAGCAGCTACACTGCTACTTGAAGTTTCCTTAATGCACCACATCGCAGATGGTCCATTCTGGCAGGTGGTCGCTGGAGACGTTGAGAATTGTAGAAGTAATTGGTGGCGTATACTCCTCCATGTACAGAATTACTACAGAAGTAATTGCATGATTCACACTTGGTATTTAGCAGTGGATTTTCAGCTCTATATTTTGTCTCCTATTGTGATTGTCTGGTTGTTCGGGAGGAAAGCGATAGCCTGGCTCATTCTGATCCTCTCAGTGTTGTTGTCATGGGCTGTGACTATAACTTACAGTTATACATTCAAGATGTCCACAATATTACATAATTTTGAGCGTTCTGCTGATATCGTTACTTATCTGGATAAGTTTTACGATAATACATTACCCAGGGCCGTACCATTCGTCGTTGGCTTGATGTTTGGGTATATCCTACACTTGCACAGAGATAAAAAGTTGACCATATCTAGGCTATATGTGTCCTTGTTCTGGATATATTCGTGCATCGTCATGTTTTTTGCGGTTTATTCTTTATACCCTATGGCCCAATCTGGCTACGATAACCAAGCGTTTGATAGTTTTTATAATGCTTTTGCTAGAATATTATGGGCTTTTATGATCGGCTGGCTAGTCTTGGCATGCGATCATGGTTATGCTGTTTACGAGTACAAAACTTGTACAAATGGCAAAAAGCAAAACGATCCTTTAAATACCAAACTGTGGAATAAAATCTACCATCATTGA
Protein
MELQGKYCTVLVPWDQMPTNLPETLEDNWLQLFLTYQFRGISGMLEQNKGYAHEVSSIRGRIDPNEHTVFVLSSNETLLGLCIPKVCTTSEALNHVLNYISFVNLTYRDYYCRLPNDKPFRPADYVSMAIFSIIAILTVASSSYEIYNVHVLKKDSSQLHPIYRNFSVFHNTRRFLTFNRNSGQLDCLDGIRVMAMSWITLGHTFCMIIFAFTHNSIYRVIKFRQLETNVIFGAILSVDTFLSISGLLLVYTVVGKIPRDRFLKGIPLFYLYRLLRIWPLLAATLLLEVSLMHHIADGPFWQVVAGDVENCRSNWWRILLHVQNYYRSNCMIHTWYLAVDFQLYILSPIVIVWLFGRKAIAWLILILSVLLSWAVTITYSYTFKMSTILHNFERSADIVTYLDKFYDNTLPRAVPFVVGLMFGYILHLHRDKKLTISRLYVSLFWIYSCIVMFFAVYSLYPMAQSGYDNQAFDSFYNAFARILWAFMIGWLVLACDHGYAVYEYKTCTNGKKQNDPLNTKLWNKIYHH
Summary
Uniprot
A0A2A4JQ97
A0A2W1BL49
A0A0L7KP15
A0A3S2NN91
A0A2W1BNT5
A0A2A4JU72
+ More
A0A3S2NE26 A0A2H1WBT1 A0A194PP47 A0A2A4JEF2 A0A212EMX9 A0A194QTR0 A0A194PIM1 A0A194QN46 A0A194QPM3 A0A3S2PE38 A0A2Z5U7W8 A0A212EMY7 A0A182P7U7 A0A182IAA6 A0A182XEE2 Q7QFJ9 A0A182TSW2 A0A182V8V1 A0A182QD03 A0A182YQ43 A0A1Q3FPF3 A0A182SBI6 Q1DH10 A0A139WIK1 A0A182R9K0 B4M0Z0 A0A182JYF2 A0A3B0K6H3 W5JPV7 B4JH76 B4NCZ2 A0A084VX41 Q179P4 A0A182W4M0 B5DL89 B3NVP9 A0A1Y1K0V2 Q16L77 A0A2J7PZI3 B4M0Z1 A0A1S4FXG2 B4HAQ3 A0A182JAD6 A0A1W7R561 B4PZG5 A0A0P6CGP4 A0A1Y1LDX2 A0A182LXU7 A0A1W4VFZ1 B5DY94 A0A2J7PZG8 A0A162QB64 A0A0P4Z2W4 B4R7R8 B4L808 A0A0P5AZ33 A0A0P4ZBN0 A0A0P4Z3D5 A0A0P4Z4S6 B3MQP9 B5DY92 B4JME3 A0A0P5S1W4 B4GB83 B4R213 A0A2M4ASR4 B4NBI1 J9KT87 A0A1Y1LRQ6 A0A1Y1LQS2 A0A3B0IZN8 Q9VCK9 A0A0N8CV45 B4PZG4 A0A0P5AVF3 A0A2J7PZF1 A0A0P5WRM4 A0A0P5E7G2 A0A0P5VTE9 A0A0P5WUR7 A0A0P5Z0C4 A0A0P5Y5T0 A0A0P5EAX5 A0A0P5VSQ1 A0A0N8A1C7 A0A0P5WPE6
A0A3S2NE26 A0A2H1WBT1 A0A194PP47 A0A2A4JEF2 A0A212EMX9 A0A194QTR0 A0A194PIM1 A0A194QN46 A0A194QPM3 A0A3S2PE38 A0A2Z5U7W8 A0A212EMY7 A0A182P7U7 A0A182IAA6 A0A182XEE2 Q7QFJ9 A0A182TSW2 A0A182V8V1 A0A182QD03 A0A182YQ43 A0A1Q3FPF3 A0A182SBI6 Q1DH10 A0A139WIK1 A0A182R9K0 B4M0Z0 A0A182JYF2 A0A3B0K6H3 W5JPV7 B4JH76 B4NCZ2 A0A084VX41 Q179P4 A0A182W4M0 B5DL89 B3NVP9 A0A1Y1K0V2 Q16L77 A0A2J7PZI3 B4M0Z1 A0A1S4FXG2 B4HAQ3 A0A182JAD6 A0A1W7R561 B4PZG5 A0A0P6CGP4 A0A1Y1LDX2 A0A182LXU7 A0A1W4VFZ1 B5DY94 A0A2J7PZG8 A0A162QB64 A0A0P4Z2W4 B4R7R8 B4L808 A0A0P5AZ33 A0A0P4ZBN0 A0A0P4Z3D5 A0A0P4Z4S6 B3MQP9 B5DY92 B4JME3 A0A0P5S1W4 B4GB83 B4R213 A0A2M4ASR4 B4NBI1 J9KT87 A0A1Y1LRQ6 A0A1Y1LQS2 A0A3B0IZN8 Q9VCK9 A0A0N8CV45 B4PZG4 A0A0P5AVF3 A0A2J7PZF1 A0A0P5WRM4 A0A0P5E7G2 A0A0P5VTE9 A0A0P5WUR7 A0A0P5Z0C4 A0A0P5Y5T0 A0A0P5EAX5 A0A0P5VSQ1 A0A0N8A1C7 A0A0P5WPE6
Pubmed
EMBL
NWSH01000795
PCG74171.1
KZ150053
PZC74364.1
JTDY01007938
KOB64845.1
+ More
RSAL01000026 RVE51984.1 KZ149975 PZC75941.1 NWSH01000663 PCG74942.1 RSAL01000079 RVE48689.1 ODYU01007612 SOQ50539.1 KQ459603 KPI92900.1 NWSH01001890 PCG69782.1 AGBW02013784 OWR42826.1 KQ461194 KPJ06931.1 KPI92898.1 KPJ06932.1 KPJ06930.1 RVE48688.1 AP017500 BBB06791.1 OWR42827.1 APCN01001136 AAAB01008846 EAA06260.5 AXCN02002156 GFDL01005570 JAV29475.1 CH899885 EAT32439.1 KQ971338 KYB27776.1 CH940650 EDW67401.2 OUUW01000005 SPP81226.1 ADMH02000869 ETN64804.1 CH916369 EDV92767.1 CH964239 EDW82701.2 ATLV01017841 KE525195 KFB42535.1 CH477347 EAT42980.1 CH379063 EDY72288.2 CH954180 EDV46714.1 GEZM01098808 GEZM01098807 JAV53750.1 CH477918 EAT35074.1 NEVH01020335 PNF21736.1 EDW67402.2 CH479242 EDW37664.1 GEHC01001326 JAV46319.1 CM000162 EDX02121.1 GDIP01000422 JAN03294.1 GEZM01058179 JAV71824.1 AXCM01007633 CM000070 EDY67570.1 PNF21712.1 LRGB01000359 KZS19531.1 GDIP01218567 JAJ04835.1 CM000366 EDX18401.1 CH933814 EDW05583.1 GDIP01192122 JAJ31280.1 GDIP01218570 JAJ04832.1 GDIP01218568 JAJ04834.1 GDIP01218571 JAJ04831.1 CH902621 EDV44675.1 EDY67568.1 CH916371 EDV92468.1 GDIQ01098087 JAL53639.1 CH479181 EDW32185.1 CM000364 EDX14070.1 GGFK01010503 MBW43824.1 CH964232 EDW81145.2 ABLF02022069 GEZM01052837 JAV74256.1 GEZM01052838 JAV74255.1 OUUW01000001 SPP73715.1 AE014297 BT058034 AAF56149.2 ACM16755.1 GDIP01095887 JAM07828.1 EDX02120.1 GDIP01197422 JAJ25980.1 PNF21711.1 GDIP01083440 JAM20275.1 GDIP01145684 JAJ77718.1 GDIP01095885 JAM07830.1 GDIP01081491 JAM22224.1 GDIP01051241 JAM52474.1 GDIP01064138 JAM39577.1 GDIP01145685 JAJ77717.1 GDIP01095886 JAM07829.1 GDIP01192121 JAJ31281.1 GDIP01085145 GDIP01083442 JAM20273.1
RSAL01000026 RVE51984.1 KZ149975 PZC75941.1 NWSH01000663 PCG74942.1 RSAL01000079 RVE48689.1 ODYU01007612 SOQ50539.1 KQ459603 KPI92900.1 NWSH01001890 PCG69782.1 AGBW02013784 OWR42826.1 KQ461194 KPJ06931.1 KPI92898.1 KPJ06932.1 KPJ06930.1 RVE48688.1 AP017500 BBB06791.1 OWR42827.1 APCN01001136 AAAB01008846 EAA06260.5 AXCN02002156 GFDL01005570 JAV29475.1 CH899885 EAT32439.1 KQ971338 KYB27776.1 CH940650 EDW67401.2 OUUW01000005 SPP81226.1 ADMH02000869 ETN64804.1 CH916369 EDV92767.1 CH964239 EDW82701.2 ATLV01017841 KE525195 KFB42535.1 CH477347 EAT42980.1 CH379063 EDY72288.2 CH954180 EDV46714.1 GEZM01098808 GEZM01098807 JAV53750.1 CH477918 EAT35074.1 NEVH01020335 PNF21736.1 EDW67402.2 CH479242 EDW37664.1 GEHC01001326 JAV46319.1 CM000162 EDX02121.1 GDIP01000422 JAN03294.1 GEZM01058179 JAV71824.1 AXCM01007633 CM000070 EDY67570.1 PNF21712.1 LRGB01000359 KZS19531.1 GDIP01218567 JAJ04835.1 CM000366 EDX18401.1 CH933814 EDW05583.1 GDIP01192122 JAJ31280.1 GDIP01218570 JAJ04832.1 GDIP01218568 JAJ04834.1 GDIP01218571 JAJ04831.1 CH902621 EDV44675.1 EDY67568.1 CH916371 EDV92468.1 GDIQ01098087 JAL53639.1 CH479181 EDW32185.1 CM000364 EDX14070.1 GGFK01010503 MBW43824.1 CH964232 EDW81145.2 ABLF02022069 GEZM01052837 JAV74256.1 GEZM01052838 JAV74255.1 OUUW01000001 SPP73715.1 AE014297 BT058034 AAF56149.2 ACM16755.1 GDIP01095887 JAM07828.1 EDX02120.1 GDIP01197422 JAJ25980.1 PNF21711.1 GDIP01083440 JAM20275.1 GDIP01145684 JAJ77718.1 GDIP01095885 JAM07830.1 GDIP01081491 JAM22224.1 GDIP01051241 JAM52474.1 GDIP01064138 JAM39577.1 GDIP01145685 JAJ77717.1 GDIP01095886 JAM07829.1 GDIP01192121 JAJ31281.1 GDIP01085145 GDIP01083442 JAM20273.1
Proteomes
UP000218220
UP000037510
UP000283053
UP000053268
UP000007151
UP000053240
+ More
UP000075885 UP000075840 UP000076407 UP000007062 UP000075902 UP000075903 UP000075886 UP000076408 UP000075901 UP000008820 UP000007266 UP000075900 UP000008792 UP000075881 UP000268350 UP000000673 UP000001070 UP000007798 UP000030765 UP000075920 UP000001819 UP000008711 UP000235965 UP000008744 UP000075880 UP000002282 UP000075883 UP000192221 UP000076858 UP000000304 UP000009192 UP000007801 UP000007819 UP000000803
UP000075885 UP000075840 UP000076407 UP000007062 UP000075902 UP000075903 UP000075886 UP000076408 UP000075901 UP000008820 UP000007266 UP000075900 UP000008792 UP000075881 UP000268350 UP000000673 UP000001070 UP000007798 UP000030765 UP000075920 UP000001819 UP000008711 UP000235965 UP000008744 UP000075880 UP000002282 UP000075883 UP000192221 UP000076858 UP000000304 UP000009192 UP000007801 UP000007819 UP000000803
PRIDE
Interpro
ProteinModelPortal
A0A2A4JQ97
A0A2W1BL49
A0A0L7KP15
A0A3S2NN91
A0A2W1BNT5
A0A2A4JU72
+ More
A0A3S2NE26 A0A2H1WBT1 A0A194PP47 A0A2A4JEF2 A0A212EMX9 A0A194QTR0 A0A194PIM1 A0A194QN46 A0A194QPM3 A0A3S2PE38 A0A2Z5U7W8 A0A212EMY7 A0A182P7U7 A0A182IAA6 A0A182XEE2 Q7QFJ9 A0A182TSW2 A0A182V8V1 A0A182QD03 A0A182YQ43 A0A1Q3FPF3 A0A182SBI6 Q1DH10 A0A139WIK1 A0A182R9K0 B4M0Z0 A0A182JYF2 A0A3B0K6H3 W5JPV7 B4JH76 B4NCZ2 A0A084VX41 Q179P4 A0A182W4M0 B5DL89 B3NVP9 A0A1Y1K0V2 Q16L77 A0A2J7PZI3 B4M0Z1 A0A1S4FXG2 B4HAQ3 A0A182JAD6 A0A1W7R561 B4PZG5 A0A0P6CGP4 A0A1Y1LDX2 A0A182LXU7 A0A1W4VFZ1 B5DY94 A0A2J7PZG8 A0A162QB64 A0A0P4Z2W4 B4R7R8 B4L808 A0A0P5AZ33 A0A0P4ZBN0 A0A0P4Z3D5 A0A0P4Z4S6 B3MQP9 B5DY92 B4JME3 A0A0P5S1W4 B4GB83 B4R213 A0A2M4ASR4 B4NBI1 J9KT87 A0A1Y1LRQ6 A0A1Y1LQS2 A0A3B0IZN8 Q9VCK9 A0A0N8CV45 B4PZG4 A0A0P5AVF3 A0A2J7PZF1 A0A0P5WRM4 A0A0P5E7G2 A0A0P5VTE9 A0A0P5WUR7 A0A0P5Z0C4 A0A0P5Y5T0 A0A0P5EAX5 A0A0P5VSQ1 A0A0N8A1C7 A0A0P5WPE6
A0A3S2NE26 A0A2H1WBT1 A0A194PP47 A0A2A4JEF2 A0A212EMX9 A0A194QTR0 A0A194PIM1 A0A194QN46 A0A194QPM3 A0A3S2PE38 A0A2Z5U7W8 A0A212EMY7 A0A182P7U7 A0A182IAA6 A0A182XEE2 Q7QFJ9 A0A182TSW2 A0A182V8V1 A0A182QD03 A0A182YQ43 A0A1Q3FPF3 A0A182SBI6 Q1DH10 A0A139WIK1 A0A182R9K0 B4M0Z0 A0A182JYF2 A0A3B0K6H3 W5JPV7 B4JH76 B4NCZ2 A0A084VX41 Q179P4 A0A182W4M0 B5DL89 B3NVP9 A0A1Y1K0V2 Q16L77 A0A2J7PZI3 B4M0Z1 A0A1S4FXG2 B4HAQ3 A0A182JAD6 A0A1W7R561 B4PZG5 A0A0P6CGP4 A0A1Y1LDX2 A0A182LXU7 A0A1W4VFZ1 B5DY94 A0A2J7PZG8 A0A162QB64 A0A0P4Z2W4 B4R7R8 B4L808 A0A0P5AZ33 A0A0P4ZBN0 A0A0P4Z3D5 A0A0P4Z4S6 B3MQP9 B5DY92 B4JME3 A0A0P5S1W4 B4GB83 B4R213 A0A2M4ASR4 B4NBI1 J9KT87 A0A1Y1LRQ6 A0A1Y1LQS2 A0A3B0IZN8 Q9VCK9 A0A0N8CV45 B4PZG4 A0A0P5AVF3 A0A2J7PZF1 A0A0P5WRM4 A0A0P5E7G2 A0A0P5VTE9 A0A0P5WUR7 A0A0P5Z0C4 A0A0P5Y5T0 A0A0P5EAX5 A0A0P5VSQ1 A0A0N8A1C7 A0A0P5WPE6
Ontologies
GO
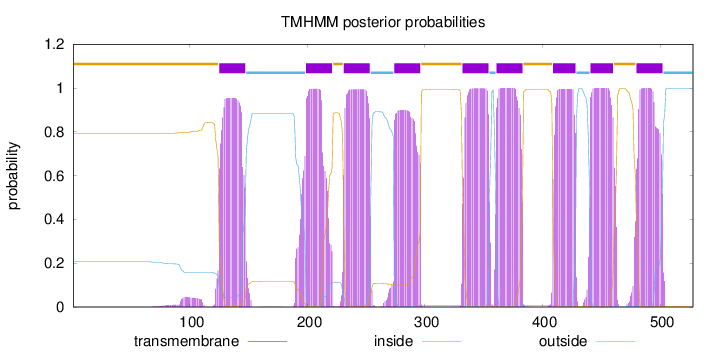
Topology
Length:
528
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
194.60375
Exp number, first 60 AAs:
0.0018
Total prob of N-in:
0.20627
outside
1 - 124
TMhelix
125 - 147
inside
148 - 198
TMhelix
199 - 221
outside
222 - 230
TMhelix
231 - 253
inside
254 - 273
TMhelix
274 - 296
outside
297 - 331
TMhelix
332 - 354
inside
355 - 360
TMhelix
361 - 383
outside
384 - 408
TMhelix
409 - 428
inside
429 - 440
TMhelix
441 - 460
outside
461 - 479
TMhelix
480 - 502
inside
503 - 528
Population Genetic Test Statistics
Pi
54.381768
Theta
172.312642
Tajima's D
-0.798234
CLR
2.02311
CSRT
0.177741112944353
Interpretation
Uncertain
