Gene
KWMTBOMO04223
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.516
Sequence
CDS
ATGCTCGGTGCTAGCACGAGCCGCCACGGGGATGTTTGCATTGAGCACGTACGTATTATAGAAAATGAAGGCGTAGGATTTGTTCATAAAAGGGTCGTTAACAACTTGCGTTATGCGGACGACACTATCCTTATTGCAGCATCGGAAGCCGATTTGCAAGCAATTGTGAATAAGGTTAATGAGTGCAGCGAAGAAGCCGGGTTGTCGATAAACATAAGCAAGACCAAGTTCATGGTAGTCTCAAGAAACCCGGATTTGAGTTCAAGCGTATCTGTAGCTGGGAAACAACTAGAGCGGGTACGACAGTACAAGTATTTAGGGGCTTGGGTAAACGAAGCATGGGAATCTGATCAAGAAATCAAGACCCGGATAGAAACTGCTCGAGCCTCTTTTAATAAAATGAGGAAAGTTCTTTGCTGTCGTCAACTGAATATAAAGCTCCGAGTCAGGCTACTCATGTGCTATATCTGGCCTATCGTCATGTATGGTTGCGAAACGTGGACCCTGAAAGAGGACACTACAAGACGCCTACAGGCATTCGAGATGTGGTGCTACCGTCGCATGTTACGCATTGGTTGGACGCAGAGGGTCAGGAACGAAACCGTGCTTCAGCGCGTTCATATGTCCCGGAAAATGCTGCCTGCTATCAAAAAGCGCAAGATAGAGTATCTCGGCCACGTGCTCAGGAACGATCGATACATGCTGCTCCAGCTGATAATTATGGGCAAAGTGGACGGAAAGAGACGTGCTGAGCGAAGAAAGAAATCATGGCTCCGGAATATCCGGGAGTGGACCGGTATTGCCTCCGTCGAACAGCTGTTCCGGTTGGCTGCAGATAAAAACGAATATAGAAAGCTAACGGCCAACCTTCAGGATTGA
Protein
MLGASTSRHGDVCIEHVRIIENEGVGFVHKRVVNNLRYADDTILIAASEADLQAIVNKVNECSEEAGLSINISKTKFMVVSRNPDLSSSVSVAGKQLERVRQYKYLGAWVNEAWESDQEIKTRIETARASFNKMRKVLCCRQLNIKLRVRLLMCYIWPIVMYGCETWTLKEDTTRRLQAFEMWCYRRMLRIGWTQRVRNETVLQRVHMSRKMLPAIKKRKIEYLGHVLRNDRYMLLQLIIMGKVDGKRRAERRKKSWLRNIREWTGIASVEQLFRLAADKNEYRKLTANLQD
Summary
Uniprot
D5LB38
A0A2A4J795
A0A146L0Z1
A0A0A9XCF5
A0A0A9XXL9
A0A0J7KRQ5
+ More
A0A0J7KZE8 A0A2S2QAF2 A0A2A4JBK9 A0A0B7BUR9 A0A0B7BVD3 A0A0B7BSZ9 A0A3S0ZE77 A0A0B7BVD9 A0A3S0ZVZ5 A0A3S1AZH6 H3B2Y3 A0A2J7PGF8 A0A2J7QV96 A0A2J7RQ44 X1XQS0 A0A2S2PJ33 A0A2J7PF33 A0A2J7R4L3 A0A2J7RFE8 A0A2J7PFP6 A0A2J7Q0H2 A0A2J7QLG4 A0A2J7RDL7 A0A2J7RAR3 A0A2J7Q7C4 A0A2J7QD50 A0A2J7RIN3 A0A2J7PGM6 A0A2J7RF43 A0A2J7QP57 A0A2J7R076 A0A2J7RSR3 A0A2J7QDT5 A0A2J7Q5G7 A0A2J7QQV1 A0A2J7QTS6 A0A2J7RKX8 A0A2J7Q6A9 A0A2J7PRL8 A0A2J7QJR1 A0A2J7RP34 A0A2J7RQF4 A0A2J7PJ38 J9KDM4 A0A2J7RIB8 A0A2J7PTP5 A0A2J7PLH7 A0A2J7R7G1 A0A2J7PL44 A0A2J7QD92 A0A2J7PJ03 A0A2J7RLC6 A0A2J7QYR3 A0A0P4W0B7 A0A2S2Q0B1 A0A2J7PZ27 A0A2J7PUJ0 A0A2J7QCS7 A0A2J7PM48 A0A2J7QCR2 A0A2J7PIT8 A0A2J7PKG5 A0A2J7PUR2 A0A2J7PXL1 A0A2J7QTX6 A0A2J7R2Z0 A0A2J7QGV0 A0A2J7PES2 A0A2J7RI73 A0A2J7QJ13 A0A2J7QSH1 A0A2J7QBX6 A0A2J7PS45 A0A2J7QTA1 A0A2J7R4G7 A0A2J7PWP4 A0A2J7Q5D5 A0A2J7RL85 A0A2J7PBA6 H2YWZ5 A0A2J7PWV5 A0A2J7Q6A8 A0A2J7R6B0 A0A0B7BT80 A0A2J7PPW0 A0A2J7R4W1 A0A2J7QPS5 A0A2J7QX53 A0A2J7R2Y9 A0A2J7Q6W4 A0A2J7R0K9
A0A0J7KZE8 A0A2S2QAF2 A0A2A4JBK9 A0A0B7BUR9 A0A0B7BVD3 A0A0B7BSZ9 A0A3S0ZE77 A0A0B7BVD9 A0A3S0ZVZ5 A0A3S1AZH6 H3B2Y3 A0A2J7PGF8 A0A2J7QV96 A0A2J7RQ44 X1XQS0 A0A2S2PJ33 A0A2J7PF33 A0A2J7R4L3 A0A2J7RFE8 A0A2J7PFP6 A0A2J7Q0H2 A0A2J7QLG4 A0A2J7RDL7 A0A2J7RAR3 A0A2J7Q7C4 A0A2J7QD50 A0A2J7RIN3 A0A2J7PGM6 A0A2J7RF43 A0A2J7QP57 A0A2J7R076 A0A2J7RSR3 A0A2J7QDT5 A0A2J7Q5G7 A0A2J7QQV1 A0A2J7QTS6 A0A2J7RKX8 A0A2J7Q6A9 A0A2J7PRL8 A0A2J7QJR1 A0A2J7RP34 A0A2J7RQF4 A0A2J7PJ38 J9KDM4 A0A2J7RIB8 A0A2J7PTP5 A0A2J7PLH7 A0A2J7R7G1 A0A2J7PL44 A0A2J7QD92 A0A2J7PJ03 A0A2J7RLC6 A0A2J7QYR3 A0A0P4W0B7 A0A2S2Q0B1 A0A2J7PZ27 A0A2J7PUJ0 A0A2J7QCS7 A0A2J7PM48 A0A2J7QCR2 A0A2J7PIT8 A0A2J7PKG5 A0A2J7PUR2 A0A2J7PXL1 A0A2J7QTX6 A0A2J7R2Z0 A0A2J7QGV0 A0A2J7PES2 A0A2J7RI73 A0A2J7QJ13 A0A2J7QSH1 A0A2J7QBX6 A0A2J7PS45 A0A2J7QTA1 A0A2J7R4G7 A0A2J7PWP4 A0A2J7Q5D5 A0A2J7RL85 A0A2J7PBA6 H2YWZ5 A0A2J7PWV5 A0A2J7Q6A8 A0A2J7R6B0 A0A0B7BT80 A0A2J7PPW0 A0A2J7R4W1 A0A2J7QPS5 A0A2J7QX53 A0A2J7R2Y9 A0A2J7Q6W4 A0A2J7R0K9
EMBL
GU815089
ADF18552.1
NWSH01002736
PCG67606.1
GDHC01017214
JAQ01415.1
+ More
GBHO01028834 JAG14770.1 GBHO01020011 GDHC01013600 JAG23593.1 JAQ05029.1 LBMM01003995 KMQ92919.1 LBMM01001656 KMQ95932.1 GGMS01004979 MBY74182.1 NWSH01001998 PCG69477.1 HACG01049251 HACG01049252 CEK96116.1 CEK96117.1 HACG01049245 CEK96110.1 HACG01049248 HACG01049253 CEK96113.1 CEK96118.1 RQTK01000625 RUS76894.1 HACG01049255 CEK96120.1 RQTK01000209 RUS84333.1 RQTK01000685 RUS76136.1 AFYH01119920 NEVH01025635 PNF15414.1 NEVH01010478 PNF32508.1 NEVH01001347 PNF42965.1 ABLF02038495 GGMR01016842 MBY29461.1 NEVH01026085 PNF14932.1 NEVH01007405 PNF35773.1 NEVH01004413 PNF39563.1 NEVH01025651 PNF15155.1 NEVH01019967 PNF22088.1 NEVH01013245 PNF29434.1 NEVH01005288 PNF38924.1 NEVH01006565 PNF37921.1 NEVH01017443 PNF24479.1 NEVH01015827 PNF26505.1 NEVH01003494 PNF40693.1 NEVH01025174 PNF15486.1 NEVH01004414 PNF39430.1 NEVH01012097 PNF30370.1 NEVH01008283 PNF34242.1 NEVH01000251 PNF43864.1 NEVH01015814 PNF26746.1 NEVH01017562 PNF23821.1 NEVH01012082 PNF30956.1 NEVH01011193 PNF31991.1 NEVH01002704 PNF41497.1 NEVH01017534 PNF24119.1 NEVH01021956 PNF18969.1 NEVH01013550 PNF28814.1 NEVH01002141 PNF42600.1 NEVH01001338 PNF43058.1 NEVH01024952 PNF16338.1 ABLF02046942 ABLF02059971 NEVH01003502 PNF40578.1 NEVH01021229 PNF19699.1 NEVH01024426 PNF17169.1 NEVH01006732 PNF36770.1 NEVH01024527 PNF17048.1 NEVH01015826 PNF26547.1 PNF16321.1 NEVH01002690 PNF41636.1 NEVH01009084 PNF33722.1 GDRN01098129 JAI59014.1 GGMS01001369 MBY70572.1 NEVH01020341 PNF21589.1 NEVH01021197 PNF19999.1 NEVH01015882 PNF26389.1 NEVH01024421 PNF17406.1 NEVH01015905 PNF26381.1 NEVH01024955 PNF16252.1 NEVH01024544 PNF16826.1 NEVH01021193 PNF20060.1 NEVH01020856 PNF21060.1 NEVH01011192 PNF32037.1 NEVH01007823 PNF35207.1 NEVH01014359 PNF27807.1 NEVH01026089 PNF14819.1 NEVH01003505 PNF40530.1 NEVH01013563 PNF28562.1 NEVH01011876 PNF31534.1 NEVH01021216 NEVH01021205 NEVH01018378 NEVH01016294 NEVH01013243 NEVH01011194 NEVH01011186 PNF19803.1 PNF19913.1 PNF23519.1 PNF26082.1 PNF29477.1 PNF31895.1 PNF32189.1 NEVH01021939 PNF19136.1 NEVH01011198 PNF31808.1 NEVH01007419 PNF35717.1 NEVH01020871 PNF20746.1 NEVH01017570 PNF23798.1 NEVH01002692 PNF41604.1 NEVH01027118 PNF13626.1 NEVH01020867 PNF20816.1 PNF24114.1 NEVH01006832 PNF36369.1 HACG01049244 HACG01049247 CEK96109.1 CEK96112.1 NEVH01022644 PNF18361.1 NEVH01007402 PNF35873.1 NEVH01012088 PNF30586.1 NEVH01009398 PNF33134.1 NEVH01007824 PNF35186.1 NEVH01017450 PNF24326.1 NEVH01008232 PNF34368.1
GBHO01028834 JAG14770.1 GBHO01020011 GDHC01013600 JAG23593.1 JAQ05029.1 LBMM01003995 KMQ92919.1 LBMM01001656 KMQ95932.1 GGMS01004979 MBY74182.1 NWSH01001998 PCG69477.1 HACG01049251 HACG01049252 CEK96116.1 CEK96117.1 HACG01049245 CEK96110.1 HACG01049248 HACG01049253 CEK96113.1 CEK96118.1 RQTK01000625 RUS76894.1 HACG01049255 CEK96120.1 RQTK01000209 RUS84333.1 RQTK01000685 RUS76136.1 AFYH01119920 NEVH01025635 PNF15414.1 NEVH01010478 PNF32508.1 NEVH01001347 PNF42965.1 ABLF02038495 GGMR01016842 MBY29461.1 NEVH01026085 PNF14932.1 NEVH01007405 PNF35773.1 NEVH01004413 PNF39563.1 NEVH01025651 PNF15155.1 NEVH01019967 PNF22088.1 NEVH01013245 PNF29434.1 NEVH01005288 PNF38924.1 NEVH01006565 PNF37921.1 NEVH01017443 PNF24479.1 NEVH01015827 PNF26505.1 NEVH01003494 PNF40693.1 NEVH01025174 PNF15486.1 NEVH01004414 PNF39430.1 NEVH01012097 PNF30370.1 NEVH01008283 PNF34242.1 NEVH01000251 PNF43864.1 NEVH01015814 PNF26746.1 NEVH01017562 PNF23821.1 NEVH01012082 PNF30956.1 NEVH01011193 PNF31991.1 NEVH01002704 PNF41497.1 NEVH01017534 PNF24119.1 NEVH01021956 PNF18969.1 NEVH01013550 PNF28814.1 NEVH01002141 PNF42600.1 NEVH01001338 PNF43058.1 NEVH01024952 PNF16338.1 ABLF02046942 ABLF02059971 NEVH01003502 PNF40578.1 NEVH01021229 PNF19699.1 NEVH01024426 PNF17169.1 NEVH01006732 PNF36770.1 NEVH01024527 PNF17048.1 NEVH01015826 PNF26547.1 PNF16321.1 NEVH01002690 PNF41636.1 NEVH01009084 PNF33722.1 GDRN01098129 JAI59014.1 GGMS01001369 MBY70572.1 NEVH01020341 PNF21589.1 NEVH01021197 PNF19999.1 NEVH01015882 PNF26389.1 NEVH01024421 PNF17406.1 NEVH01015905 PNF26381.1 NEVH01024955 PNF16252.1 NEVH01024544 PNF16826.1 NEVH01021193 PNF20060.1 NEVH01020856 PNF21060.1 NEVH01011192 PNF32037.1 NEVH01007823 PNF35207.1 NEVH01014359 PNF27807.1 NEVH01026089 PNF14819.1 NEVH01003505 PNF40530.1 NEVH01013563 PNF28562.1 NEVH01011876 PNF31534.1 NEVH01021216 NEVH01021205 NEVH01018378 NEVH01016294 NEVH01013243 NEVH01011194 NEVH01011186 PNF19803.1 PNF19913.1 PNF23519.1 PNF26082.1 PNF29477.1 PNF31895.1 PNF32189.1 NEVH01021939 PNF19136.1 NEVH01011198 PNF31808.1 NEVH01007419 PNF35717.1 NEVH01020871 PNF20746.1 NEVH01017570 PNF23798.1 NEVH01002692 PNF41604.1 NEVH01027118 PNF13626.1 NEVH01020867 PNF20816.1 PNF24114.1 NEVH01006832 PNF36369.1 HACG01049244 HACG01049247 CEK96109.1 CEK96112.1 NEVH01022644 PNF18361.1 NEVH01007402 PNF35873.1 NEVH01012088 PNF30586.1 NEVH01009398 PNF33134.1 NEVH01007824 PNF35186.1 NEVH01017450 PNF24326.1 NEVH01008232 PNF34368.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
D5LB38
A0A2A4J795
A0A146L0Z1
A0A0A9XCF5
A0A0A9XXL9
A0A0J7KRQ5
+ More
A0A0J7KZE8 A0A2S2QAF2 A0A2A4JBK9 A0A0B7BUR9 A0A0B7BVD3 A0A0B7BSZ9 A0A3S0ZE77 A0A0B7BVD9 A0A3S0ZVZ5 A0A3S1AZH6 H3B2Y3 A0A2J7PGF8 A0A2J7QV96 A0A2J7RQ44 X1XQS0 A0A2S2PJ33 A0A2J7PF33 A0A2J7R4L3 A0A2J7RFE8 A0A2J7PFP6 A0A2J7Q0H2 A0A2J7QLG4 A0A2J7RDL7 A0A2J7RAR3 A0A2J7Q7C4 A0A2J7QD50 A0A2J7RIN3 A0A2J7PGM6 A0A2J7RF43 A0A2J7QP57 A0A2J7R076 A0A2J7RSR3 A0A2J7QDT5 A0A2J7Q5G7 A0A2J7QQV1 A0A2J7QTS6 A0A2J7RKX8 A0A2J7Q6A9 A0A2J7PRL8 A0A2J7QJR1 A0A2J7RP34 A0A2J7RQF4 A0A2J7PJ38 J9KDM4 A0A2J7RIB8 A0A2J7PTP5 A0A2J7PLH7 A0A2J7R7G1 A0A2J7PL44 A0A2J7QD92 A0A2J7PJ03 A0A2J7RLC6 A0A2J7QYR3 A0A0P4W0B7 A0A2S2Q0B1 A0A2J7PZ27 A0A2J7PUJ0 A0A2J7QCS7 A0A2J7PM48 A0A2J7QCR2 A0A2J7PIT8 A0A2J7PKG5 A0A2J7PUR2 A0A2J7PXL1 A0A2J7QTX6 A0A2J7R2Z0 A0A2J7QGV0 A0A2J7PES2 A0A2J7RI73 A0A2J7QJ13 A0A2J7QSH1 A0A2J7QBX6 A0A2J7PS45 A0A2J7QTA1 A0A2J7R4G7 A0A2J7PWP4 A0A2J7Q5D5 A0A2J7RL85 A0A2J7PBA6 H2YWZ5 A0A2J7PWV5 A0A2J7Q6A8 A0A2J7R6B0 A0A0B7BT80 A0A2J7PPW0 A0A2J7R4W1 A0A2J7QPS5 A0A2J7QX53 A0A2J7R2Y9 A0A2J7Q6W4 A0A2J7R0K9
A0A0J7KZE8 A0A2S2QAF2 A0A2A4JBK9 A0A0B7BUR9 A0A0B7BVD3 A0A0B7BSZ9 A0A3S0ZE77 A0A0B7BVD9 A0A3S0ZVZ5 A0A3S1AZH6 H3B2Y3 A0A2J7PGF8 A0A2J7QV96 A0A2J7RQ44 X1XQS0 A0A2S2PJ33 A0A2J7PF33 A0A2J7R4L3 A0A2J7RFE8 A0A2J7PFP6 A0A2J7Q0H2 A0A2J7QLG4 A0A2J7RDL7 A0A2J7RAR3 A0A2J7Q7C4 A0A2J7QD50 A0A2J7RIN3 A0A2J7PGM6 A0A2J7RF43 A0A2J7QP57 A0A2J7R076 A0A2J7RSR3 A0A2J7QDT5 A0A2J7Q5G7 A0A2J7QQV1 A0A2J7QTS6 A0A2J7RKX8 A0A2J7Q6A9 A0A2J7PRL8 A0A2J7QJR1 A0A2J7RP34 A0A2J7RQF4 A0A2J7PJ38 J9KDM4 A0A2J7RIB8 A0A2J7PTP5 A0A2J7PLH7 A0A2J7R7G1 A0A2J7PL44 A0A2J7QD92 A0A2J7PJ03 A0A2J7RLC6 A0A2J7QYR3 A0A0P4W0B7 A0A2S2Q0B1 A0A2J7PZ27 A0A2J7PUJ0 A0A2J7QCS7 A0A2J7PM48 A0A2J7QCR2 A0A2J7PIT8 A0A2J7PKG5 A0A2J7PUR2 A0A2J7PXL1 A0A2J7QTX6 A0A2J7R2Z0 A0A2J7QGV0 A0A2J7PES2 A0A2J7RI73 A0A2J7QJ13 A0A2J7QSH1 A0A2J7QBX6 A0A2J7PS45 A0A2J7QTA1 A0A2J7R4G7 A0A2J7PWP4 A0A2J7Q5D5 A0A2J7RL85 A0A2J7PBA6 H2YWZ5 A0A2J7PWV5 A0A2J7Q6A8 A0A2J7R6B0 A0A0B7BT80 A0A2J7PPW0 A0A2J7R4W1 A0A2J7QPS5 A0A2J7QX53 A0A2J7R2Y9 A0A2J7Q6W4 A0A2J7R0K9
PDB
5HHL
E-value=0.0250114,
Score=86
Ontologies
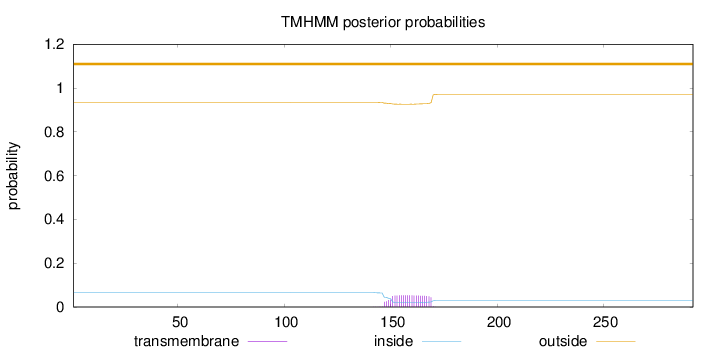
Topology
Length:
292
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.1003
Exp number, first 60 AAs:
0
Total prob of N-in:
0.06562
outside
1 - 292
Population Genetic Test Statistics
Pi
4.436102
Theta
15.772722
Tajima's D
-1.408825
CLR
16.67861
CSRT
0.0686965651717414
Interpretation
Uncertain
